Abstract
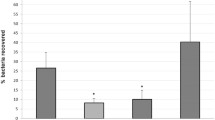
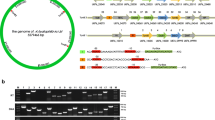
Aeromonas salmonicida subsp. salmonicida, the causative agent of furunculosis in fish, can use heme as the sole iron source. We applied the Fur Titration Assay to isolate a cluster including six genes hutAZXBCD that showed similarity to heme uptake genes of other Gram-negative bacteria, and three genes orf123 of unknown function. The spatial organization of these nine genes, arranged in five transcriptional units, was similar to that of a homologous cluster in A. hydrophila. When a TonB system was provided, this cluster allowed Escherichia coli 101ESD (an ent mutant, unable to synthesize enterobactin) to utilize hemin and hemoglobin as iron sources. Mutation of hutB, a gene that encodes a predicted periplasmic hemin-binding protein, caused a drastic defect in the ability of A. salmonicida to grow with hemin as unique source of iron. Interestingly, a mutant for hutA gene (encoding the outer membrane hemin receptor) showed initially a reduced ability to grow with hemin as sole iron source, but after 24 h it achieved growth levels similar to parental strain. Thus mutation of hutA could not abolish the growth with hemin as iron source, suggesting that redundant outer membrane heme transport functions might be encoded in the A. salmonicida genome.



Similar content being viewed by others
References
Braun V, Hantke K, Köster W (1998) Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst 35:67–145
Chart H, Trust TJ (1983) Acquisition of iron by Aeromonas salmonicida. J Bacteriol 156:758–764
Correa N, Lauriano CM, McGee R, Klose KE (2000) Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol 35:743–755
Crosa JH, Walsh CT (2002) Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249
de Lorenzo V, Wee S, Herrero M, Neilands JB (1987) Operator sequence of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169:2624–2630
Ebanks RO, Dacanay A, Goguen M, Pinto DM, Ross NW (2004) Differential proteomic analysis of Aeromonas salmonicida outer membrane proteins in response to low iron and in vivo growth conditions. Proteomics 4:1074–1085
Fernandez AIG, Fernández AF, Pérez MJ, Nieto TP, Ellis AE (1998) Siderophore production by Aeromonas salmonicida subsp. salmonicida. Lack of strain specificity. Dis Aquat Organ 33:87–97
Genco CA, Dixon DW (2001) Emerging strategies in microbial haem capture. Mol Microbiol 39:1–11
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hantke K (1987) Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K12: fur not only affects iron metabolism. Mol Gen Genet 210:135–139
Henderson DP, Payne SM (1993) Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol Microbiol 7:461–469
Henderson DP, Payne SM (1994) Vibrio cholerae iron transport systems: role of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun 62:5120–5125
Henderson DP, Wyckoff EE, Rashidi CE, Oldham AL (2001) Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J Bacteriol 183:2715–2723
Herrero M, de Lorenzo V, Timmis KN (1990) Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J Bacteriol 172:6557–6567
Hirst ID, Hasting TS, Ellis AE (1991) Siderophore production by Aeromonas salmonicida. J Gen Microbiol 137:1185–1192
Ishiguro EE, Ainsworth T, Kay WW, Trust TJ (1986) Heme requirement for growth of fastidious atypical strains of Aeromonas salmonicida. Appl Environ Microbiol 51:668–670
Köster W (2001) ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res Microbiol 152:291–301
Mazoy R, Osorio CR, Toranzo AE, Lemos ML (2003) Isolation of mutants of Vibrio anguillarum defective in haeme utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch Microbiol 179:329–338
Mey AR, Payne SM (2001) Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol 42:835–849
Mills M, Payne SM (1997) Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect Immun 65:5358–5363
Mouriño S, Osorio CR, Lemos ML (2004) Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J Bacteriol 186:6159–6167
Mouriño S, Rodríguez-Ares I, Osorio CR, Lemos ML (2005) Genetic variability of the heme uptake system among different strains of the fish pathogen Vibrio anguillarum: identification of a new heme receptor. Appl Environ Microbiol 71:8434–8441
Najimi M, Lemos ML, Osorio CR (2008) Identification of siderophore biosynthesis genes essential for growth under iron limitation in Aeromonas salmonicida. Appl Environ Microbiol 74:2341–2348
Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM (1998) Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol 29:1493–1507
Osorio CR, Lemos ML (2002) Haeme iron acquisition mechanisms in Vibrionaceae. In: Pandalai SG (ed) Recent research developments in microbiology, vol 6. Research Signpost, Trivandrum, pp 419–436
Panek H, O’Brian MR (2002) A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273–2282
Ratledge C, Dover LG (2000) Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54:881–941
Reidl J, Mekalanos JJ (1996) Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med 183:621–629
Rio SJ, Osorio CR, Lemos ML (2005) Heme uptake genes in human and fish isolates of Photobacterium damselae: existence of hutA pseudogenes. Arch Microbiol 183:347–358
Rose RE (1988) The nucleotide sequence of pACYC177. Nucleic Acids Res 16:356
Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF (2006) Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol 188:8272–8282
Stojiljkovic I, Hantke K (1992) Hemin uptake system of Yersinia enterocolitica: similarities with the other TonB-dependent systems in Gram-negative bacteria. EMBO J 11:4359–4367
Stojiljkovic I, Bäumler AJ, Hantke K (1994) Fur regulon in Gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol 236:531–545
Stojiljkovic I, Hwa V, de Saint Martin L, O’Gaora P, Nassif X, Heffron F, So M (1995) The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol 15:531–541
Tabor S, Richardson CC (1985) A bacteriophage T7 RNA polymerase-promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA 82:1074–1078
Toranzo AE, Magariños B, Romalde JL (2005) A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246:37–61
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951
Wang RF, Kushner SR (1991) Construction of versatile low-copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene 100:195–199
Wyckoff EE, Schmitt M, Wilks A, Payne SM (2004) HutZ is required for efficient heme utilization in Vibrio cholerae. J Bacteriol 186:4142–4151
Acknowledgments
This work was supported by Grant PGIDIT06RMA26101PR-2 and Contract No. 2004/CP481, from Xunta de Galicia, Spain. Mohsen Najimi acknowledges the Ministry of Science and Education of Iran for a predoctoral fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jorge Membrillo-Hernández.
Rights and permissions
About this article
Cite this article
Najimi, M., Lemos, M.L. & Osorio, C.R. Identification of heme uptake genes in the fish pathogen Aeromonas salmonicida subsp. salmonicida . Arch Microbiol 190, 439–449 (2008). https://doi.org/10.1007/s00203-008-0391-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-008-0391-5