Abstract
Purpose
Various studies have analyzed sepsis subtypes, yet the reproducibility of such results remains unclear. This study aimed to determine the reproducibility of sepsis subtypes across multiple cohorts.
Methods
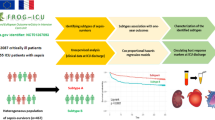
The study examined 63,547 sepsis patients from six distinct cohorts who had similar sepsis-related characteristics (vital signs, lactate, sequential organ failure assessment score, bilirubin, serum, urine output, and Glasgow coma scale). Identical cluster analysis techniques were used, employing 27 clustering schemes, and normalized mutual information (NMI), a metric ranging from 0 to 1 with higher values indicating better concordance, was employed to quantify the clustering solutions' reproducibility. Principal component analysis (PCA) was utilized to obtain the disease axis, and its uniformity across cohorts was evaluated through patterns of feature loading and correlation.
Results
The reproducibility of sepsis clustering subtypes across the various studies was modest (median NMI ranging from 0.08 to 0.54). The top-down transfer learning method (model trained on cohorts with greater severity was transferred to cohorts with lower severity score) had a higher NMI value than the bottom-up approach (median [Q1, Q3]: 0.64 [0.49, 0.78] vs. 0.23 [0.2, 0.31], p < 0.001). The reproducibility was greater when the transfer solution was performed within United States (US) cohorts. The PCA analysis revealed that the correlation pattern between variables was consistent across all cohorts, and the first two disease axes were the "shock axis" and "systemic inflammatory response syndrome (SIRS) axis."
Conclusions
Cluster analysis of sepsis patients across various cohorts showed modest reproducibility. Sepsis heterogeneity is better characterized through continuous disease axes that coexist to varying degrees within the same individual instead of mutually exclusive subtypes.




Similar content being viewed by others
Data availability
Datasets can be obtained by contacting the corresponding author upon a reasonable request.
References
Rajendran S, Xu Z, Pan W, Ghosh A, Wang F (2023) Data heterogeneity in federated learning with electronic health records: case studies of risk prediction for acute kidney injury and sepsis diseases in critical care. PLOS Digit Health 2:e0000117
Wang W, Liu C-F (2023) Sepsis heterogeneity. World J Pediatr. https://doi.org/10.1007/s12519-023-00689-8
Harhay MO, Casey JD, Clement M et al (2020) Contemporary strategies to improve clinical trial design for critical care research: insights from the first critical care clinical trialists workshop. Intensive Care Med 46:930–942
François B, Lambden S, Fivez T et al (2023) Prospective evaluation of the efficacy, safety, and optimal biomarker enrichment strategy for nangibotide, a TREM-1 inhibitor, in patients with septic shock (ASTONISH): a double-blind, randomised, controlled, phase 2b trial. Lancet Respir Med. https://doi.org/10.1016/S2213-2600(23)00158-3
Reignier J, Plantefeve G, Mira J-P et al (2023) Low versus standard calorie and protein feeding in ventilated adults with shock: a randomised, controlled, multicentre, open-label, parallel-group trial (NUTRIREA-3). Lancet Respir Med. https://doi.org/10.1016/S2213-2600(23)00092-9
Zhang Z, Zhang G, Goyal H, Mo L, Hong Y (2018) Identification of subclasses of sepsis that showed different clinical outcomes and responses to amount of fluid resuscitation: a latent profile analysis. Crit Care 22:347
Seymour CW, Kennedy JN, Wang S et al (2019) Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA 321:2003–2017
Mosevoll KA, Hansen BA, Gundersen IM et al (2023) Systemic metabolomic profiles in adult patients with bacterial sepsis: characterization of patient heterogeneity at the time of diagnosis. Biomolecules 13:223
Qin Y, Caldino Bohn RI, Sriram A et al (2023) Refining empiric subgroups of pediatric sepsis using machine-learning techniques on observational data. Front Pediatr 11:1035576
Charrad M, Ghazzali N, Boiteau V, Niknafs A (2014) NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Soft 61:1–36
Kinney GL, Santorico SA, Young KA et al (2018) Identification of chronic obstructive pulmonary disease axes that predict all-cause mortality: the COPDGene study. Am J Epidemiol 187:2109–2116
Thoral PJ, Peppink JM, Driessen RH et al (2021) Sharing ICU patient data responsibly under the society of critical care medicine/European society of intensive care medicine joint data science collaboration: the Amsterdam university medical centers database (AmsterdamUMCdb) example. Crit Care Med 49:e563–e577
Zhang Z, Chen L, Liu H et al (2022) Gene signature for the prediction of the trajectories of sepsis-induced acute kidney injury. Crit Care 26:398
Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O (2018) The eICU collaborative research database, a freely available multi-center database for critical care research. Sci Data 5:180178
Xu P, Chen L, Zhu Y et al (2022) Critical care database comprising patients with infection. Front Public Health 10:852410
Johnson AEW, Bulgarelli L, Shen L et al (2023) MIMIC-IV, a freely accessible electronic health record dataset. Sci Data 10:1
Singer M, Deutschman CS, Seymour CW et al (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315:801–810
Sterne JAC, White IR, Carlin JB et al (2009) Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ 338:b2393
Vesin A, Azoulay E, Ruckly S et al (2013) Reporting and handling missing values in clinical studies in intensive care units. Intensive Care Med 39:1396–1404
Djouzi K, Beghdad-Bey K (2019) A review of clustering algorithms for big data. In: 2019 International Conference on Networking and Advanced Systems (ICNAS). pp. 1–6
Cervantes J, Garcia-Lamont F, Rodríguez-Mazahua L, Lopez A (2020) A comprehensive survey on support vector machine classification: applications, challenges and trends. Neurocomputing 408:189–215
Vinh NX, Epps J, Bailey J (2009) Information theoretic measures for clusterings comparison: is a correction for chance necessary? In: Proceedings of the 26th Annual International Conference on Machine Learning. New York, NY, USA: Association for Computing Machinery, pp. 1073–80
Lever J, Krzywinski M, Altman N (2017) Principal component analysis. Nat Methods 14:641–642
Bhavani SV, Semler M, Qian ET et al (2022) Development and validation of novel sepsis subphenotypes using trajectories of vital signs. Intensive Care Med 48:1582–1592
Rogers AJ, Leligdowicz A, Contrepois K et al (2021) Plasma metabolites in early sepsis identify distinct clusters defined by plasma lipids. Crit Care Explor 3:e0478
Castaldi PJ, Benet M, Petersen H et al (2017) Do “COPD subtypes” really exist? Thorax 72:998–1006
Russell JA, Gordon AC, Williams MD, Boyd JH, Walley KR, Kissoon N (2021) Vasopressor therapy in the intensive care unit. Semin Respir Crit Care Med 42:59–77
Li Y, Sun P, Chang K et al (2022) Effect of continuous renal replacement therapy with the oXiris hemofilter on critically ill patients: a narrative review. J Clin Med 11:6719
Funding
ZZ received funding from the Open Foundation of Key Laboratory of Digital Technology in Medical Diagnostics of Zhejiang Province (SZZD202206), Health Science and Technology Plan of Zhejiang Province (2021KY745), the Fundamental Research Funds for the Central Universities (226–2022-00148), National natural science foundation of China (82272180) and the Project of Drug Clinical Evaluate Research of Chinese Pharmaceutical Association NO.CPA-Z06-ZC-2021–004. YH received funding from the Key Research & Development project of Zhejiang Province (2021C03071).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Z., Chen, L., Liu, X. et al. Exploring disease axes as an alternative to distinct clusters for characterizing sepsis heterogeneity. Intensive Care Med 49, 1349–1359 (2023). https://doi.org/10.1007/s00134-023-07226-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-023-07226-1