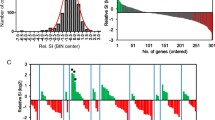
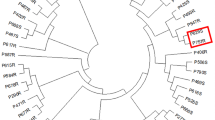
Abstract
Xanthomonas translucens pv. graminis (Xtg) causes bacterial wilt, a severe disease of forage grasses such as Italian ryegrass (Lolium multiflorum Lam.). In order to gain a more detailed understanding of the genetic control of resistance mechanisms and to provide prerequisites for marker assisted selection, the partial transcriptomes of two Italian ryegrass genotypes, one resistant and one susceptible to bacterial wilt were compared at four time points after Xtg infection. A cDNA microarray developed from a perennial ryegrass (Lolium perenne) expressed sequence tag set consisting of 9,990 unique genes was used for transcriptome analysis in Italian ryegrass. An average of 4,487 (45%) of the perennial ryegrass sequences spotted on the cDNA microarray were detected by cross-hybridisation to Italian ryegrass. Transcriptome analyses of the resistant versus the susceptible genotype revealed substantial gene expression differences (>1,200) indicating that great gene expression differences between different Italian ryegrass genotypes exist which potentially contribute to the observed phenotypic divergence in Xtg resistance between the two genotypes. In the resistant genotype, several genes differentially expressed after Xtg inoculation were identified which revealed similarities to transcriptional changes triggered by pathogen-associated molecular patterns in other plant–pathogen interactions. These genes represent candidate genes of particular interest for the development of tools for marker assisted resistance breeding.






Similar content being viewed by others
References
An QL, Ehlers K, Kogel KH, van Bel AJE, Huckelhoven R (2006) Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol 172:563–576
Andersen J, Jensen L, Asp T, Lübberstedt T (2006) Vernalization response in perennial ryegrass (Lolium perenne L.) involves orthologues of diploid wheat (Triticum monococcum) VRN1 and rice (Oryza sativa) Hd1. Plant Mol Biol 60:481
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Asp T, Frei UK, Didion T, Nielsen KK, Lubberstedt T (2007) Frequency, type, and distribution of EST-SSRs from three genotypes of Lolium perenne, and their conservation across orthologous sequences of Festuca arundinacea, Brachypodium distachyon, and Oryza sativa. BMC Plant Biol 7:36
Balaji V, Gibly A, Debbie P, Sessa G (2007) Transcriptional analysis of the tomato resistance response triggered by recognition of the Xanthomonas type III effector AvrXv3. Funct Integr Genomics 7:305–316
Bar-Or C, Czosnek H, Koltai H (2007) Cross-species microarray hybridisations: a developing tool for studying species diversity. Trends Genet 23:200–207
Becher M, Talke IN, Krall L, Kramer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37:251–268
Bernier F, Berna A (2001) Germins and germin-like proteins: plant do-all proteins. But what do they do exactly? Plant Physiol Biochem 39:545–554
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalisation methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Cao H, Glazebrook J, Clarke JD, Volko S, Dong XN (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88:57–63
Catalan P, Torrecilla P, Rodriguez JAL, Olmstead RG (2004) Phylogeny of the festucoid grasses of subtribe Loliinae and allies (Poeae, Pooideae) inferred from ITS and trnL-F sequences. Mol Phylogenet Evol 31:517–541
Cernadas RA, Camillo LR, Benedetti CE (2008) Transcriptional analysis of the sweet orange interaction with the citrus canker pathogens Xanthomonas axonopodis pv. citri and Xanthomonas axonopodis pv. aurantifolii. Mol Plant Pathol 9:609–631
Chen XW, Hackett CA, Niks RE, Hedley PE, Booth C, Druka A, Marcel TC, Vels A, Bayer M, Milne I, Morris J, Ramsay L, Marshall D, Cardle L, Waugh R (2010) An eQTL analysis of partial resistance to Puccinia hordei in barley. Plos One 5(1):e8598
Collins A, Milbourne D, Ramsay L, Meyer R, Chatot-Balandras C, Oberhagemann P, De Jong W, Gebhardt C, Bonnel E, Waugh R (1999) QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol Breed 5:387–398
Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Huckelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425:973–977
Devos KM (2005) Updating the ‘crop circle’. Curr Opin Plant Biol 8:155–162
Dracatos PM, Cogan NOI, Sawbridge TI, Gendall AR, Smith KF, Spangenberg GC, Forster JW (2009) Molecular characterisation and genetic mapping of candidate genes for qualitative disease resistance in perennial ryegrass (Lolium perenne L.). BMC Plant Biol 9:62
Egli T, Schmidt D (1982) Pathogenic variation among the causal agents of bacterial wilt of forage grasses. Phytopathol Z 104:138–150
Egli T, Goto M, Schmidt D (1975) Bacterial wilt, a new forage grass disease. Phytopathol Z 82:111–121
Gibly A, Bonshtien A, Balaji V, Debbie P, Martin GB, Sessa G (2004) Identification and expression profiling of tomato genes differentially regulated during a resistance response to Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 17:1212–1222
Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP (2006) Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440:242–245
Gregersen PL, Brinch-Pedersen H, Holm PB (2005) A microarray-based comparative analysis of gene expression profiles during grain development in transgenic and wild type wheat. Transgenic Res 14:887–905
Ikeda S (2005) Isolation of disease resistance gene analogs from Italian ryegrass (Lolium multiflorum Lam.). Grassl Sci 51:63–70
Inoue M, Gao Z, Hirata M, Fujimori M, Cai H (2004) Construction of a high-density linkage map of Italian ryegrass (Lolium multiflorum Lam.) using restriction fragment length polymorphism, amplified fragment length polymorphism, and telomeric repeat associated sequence markers. Genome 47:57–65
Kay S, Bonas U (2009) How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol 12:37–43
Kölliker R, Kraehenbuehl R, Boller B, Widmer F (2006) Genetic diversity and pathogenicity of the grass pathogen Xanthomonas translucens pv. graminis. Syst Appl Microbiol 29:109–119
Kottapalli KR, Satoh K, Rakwal R, Shibato J, Doi K, Nagata T, Kikuchi S (2007) Combining in silico mapping and arraying: an approach to identifying common candidate genes for submergence tolerance and resistance to bacterial leaf blight in ice. Mol Cells 24:394–408
Li Q, Chen F, Sun LX, Zhang ZQ, Yang YN, He ZH (2006) Expression profiling of rice genes in early defense responses to blast and bacterial blight pathogens using cDNA microarray. Physiol Mol Plant Pathol 68:51–60
Lonnstedt I, Speed T (2002) Replicated microarray data. Stat Sin 12:31–46
Ma JF, Yamaji N (2006) Silicon uptake and accumulation in higher plants. Trends Plant Sci 11:392–397
Manosalva PM, Davidson RM, Liu B, Zhu XY, Hulbert SH, Leung H, Leach JE (2009) A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol 149:286–296
Martin RC, Hollenbeck VG, Dombrowski JE (2008) Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Sci 48:1881–1887
Michel VV (2001) Interactions between Xanthomonas campestris pv. graminis strains and meadow fescue and Italian rye grass cultivars. Plant Dis 85:538–542
Moore S, Payton P, Wright M, Tanksley S, Giovannoni J (2005) Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot 56:2885–2895
Paquet A, Yang JYH (2008) arrayQuality: assessing array quality on spotted arrays, R package version 1.24.0. http://arrays.ucsf.edu/
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30(9):e36
Posselt U (2010) Breeding methods in cross-pollinated species. In: Boller B, Posselt U, Veronesi F (eds) Handbook of plant breeding: fodder crops and amenity grasses. Springer, New York, pp 39–88
Qin LX, Kerr KF (2004) Empirical evaluation of data transformations and ranking statistics for microarray analysis. Nucleic Acids Res 32:5471–5479
Ramalingam J, Cruz CMV, Kukreja K, Chittoor JM, Wu JL, Lee SW, Baraoidan M, George ML, Cohen MB, Hulbert SH, Leach JE, Leung H (2003) Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact 16:14–24
Rechsteiner MP, Widmer F, Kölliker R (2006) Expression profiling of Italian ryegrass (Lolium multiflorum Lam.) during infection with the bacterial wilt inducing pathogen Xanthomonas translucens pv. graminis. Plant Breed 125:43–51
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277:10555–10561
Schejbel B, Jensen LB, Asp T, Xing Y, Lubberstedt T (2008) Mapping of QTL for resistance to powdery mildew and resistance gene analogues in perennial ryegrass. Plant Breed 127:368–375
Seong ES, Choi D, Cho HS, Lim CK, Cho HJ, Wang MH (2007) Characterisation of a stress-responsive ankyrin repeat-containing zinc finger protein of Capsicum annuum (CaKR1). J Biochem Mol Biol 40:952–958
Silva IT, Rodrigues FA, Oliveira JR, Pereira SC, Andrade CCL, Silveira PR, Conciecao MM (2010) Wheat resistance to bacterial leaf streak mediated by silicon. J Phytopathol 158:253–262
Smyth GK (2005) Limma: linear models for microarray data. Springer, New York
Studer B, Boller B, Herrmann D, Bauer E, Posselt UK, Widmer F, Kölliker R (2006) Genetic mapping reveals a single major QTL for bacterial wilt resistance in Italian ryegrass (Lolium multiflorum Lam.). Theor Appl Genet 113:661–671
Studer B, Kölliker R, Muylle H, Asp T, Frei U, Roldán-Ruiz I, Barre P, Tomaszewski C, Meally H, Barth S, Skøt L, Armstead IP, Dolstra O, Lübberstedt T (2010) EST-derived SSR markers used as anchor loci for the construction of a consensus linkage map in ryegrass (Lolium spp.). BMC Plant Biol 10:177
Valasek MA, Repa JJ (2005) The power of real-time PCR. Adv Physiol Educ 29:151–159
Vauterin L, Hoste B, Kersters K, Swings J (1995) Reclassification of Xanthomonas. Int J Syst Bacteriol 45:472–489
Wang HM, Sletten A (1995) Infection biology of bacterial wilt of forage grasses. J Phytopathol 143:141–145
Weber M, Harada E, Vess C, von Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37:269–281
Wichmann F, Müller-Hug B, Widmer F, Boller B, Studer B, Kölliker R (2010) Phenotypic and molecular genetic characterisation indicate no major race-specific interactions between Xanthomonas translucens pv. graminis and Lolium multiflorum. Plant Pathol. doi: 10.1111/j.1365-3059.2010.02373.x
Wright SY, Suner MM, Bell PJ, Vaudin M, Greenland AJ (1993) Isolation and characterisation of male flower cDNAs from maize. Plant J 3:41–49
Yang YH, Speed T (2002) Design issues for cDNA microarray experiments. Nat Rev Genet 3:579–588
Acknowledgments
This research was funded by the Swiss National Science Foundation SNF Project (3100A0-112582). We would like to thank L. B. Jensen, B. Studer and L. B. Holte for technical and personal assistance in the department of Genetics and Biotechnology at Det Jorbrugvidenskabelige Fakultet (DJF), University of Aarhus. Special thanks are extended to S. Reinhard and P. Streckeisen for technical assistance at Agroscope Reckenholz-Tänikon, H.-P. Piepho and A. Schützenmeister for help with the experimental design and H. Rehrauer from the Functional Genomics Center Zürich for assistance with the statistical analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Luebberstedt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wichmann, F., Asp, T., Widmer, F. et al. Transcriptional responses of Italian ryegrass during interaction with Xanthomonas translucens pv. graminis reveal novel candidate genes for bacterial wilt resistance. Theor Appl Genet 122, 567–579 (2011). https://doi.org/10.1007/s00122-010-1470-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1470-y