Abstract
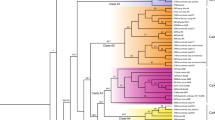
Cytoplasmically inherited characters such as resistance to viral and fungal diseases, determination of starch types, crop yield, resistance to low or high temperature often contribute to the advantageous phenotypic traits of plants. In the present study, our goal was to elucidate the genealogy of cytoplasmic genomes chloroplast and mitochondria in banana. Banana breeding is rather complicated because of the low fertility and mostly unknown origin of the edible cultivars, therefore, knowledge on the putative fertile ancestors of cytoplasmic genomes chloroplast and mitochondria would be beneficial for breeding programmes. Based on the established marker systems distinct species specific gene-pools could be identified for both chloroplast and mitochondrial genomes for Musa acuminata and Musa balbisiana wild types, respectively. Detailed analysis of the species specific chloroplast and mitochondrial gene-pools of M. acuminata and M. balbisiana revealed six chloroplast and seven mitochondrial gene-pools in the analysed accessions. Comparative analysis of the haplotypes revealed the presence of Primary Centers of origin for both chloroplast and mitochondrial genomes of both species supporting the idea of common origin of these genomes. Cytotypes representing combinations of M. acuminata chloroplast and mitochondrial gene-pools were identified in majority of the analysed hybrid cultivars. A single M. acuminata cytotype was present in the majority of the analysed cultivars, which combination was not detected in any of the wild types. On the other part a single balbisiana cytotype was identified participating in the formation of interspecies hybrids. The strong preference for the presence of certain cytoplasmic gene-pools in cultivars may indicate hundreds of years of natural as well as of farmers’ selection supplementing the phenotypic traits provided by the nuclear genome. Based on the present results the present day subspecies classification of M. acuminata is also discussed.





Similar content being viewed by others
References
Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13:1803–1818
Braun CJ, Siedow JN, Williams ME, Levings CS (1989) Mutations in the maize mitochondrial T-urf13 gene eliminate sensitivity to a fungal pathotoxin. Proc Natl Acad Sci USA 86:4435–4439
Carreel F, Gonzalez de Leon D, Lagoda P et al (2002) Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45:679–692
Cheesman EE (1950) The classification of the bananas. Kew Bull 5, pp 27–31, 151–155
Chen J, Zhu J (1999) Genetic effects and genotype × environment interactions for cooking quality traits in Indica-japonica crosses of rice (Oryza sativa L.). Euphytica 109:9–15
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Duminil J, Pemonge M-H, Petit RJ (2002) A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA. Mol Ecol Notes 2:428–430
Dumolin-Lapègue S, Demesure B, Le Corre V, Fineschi S, Petit RJ (1997) Phylogeographic structure of white oaks throughout the European continent. Genetics 146:1475–1487
Ekiz H, Konzak CF (1991) Nuclear and cytoplasmic control of anther culture response in wheat I. Analyses of alloplasmic lines. Crop Sci 31:1421–1427
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Faure S, Noyer JL, Carreel F, Horry JP, Bakry F, Lanaud C (1993) Maternal inheritance of chloroplast genome and paternal inheritance of mitochondrial genome in bananas (Musa acuminata). Curr Genet 25:265–269
Ge XJ, Liu MH, Wang WK, Schaal BA, Chiang TY (2005) Population structure of wild bananas, Musa balbisiana, in China determined by SSR fingerprinting and cpDNA PCR-RFLP. Mol Ecol 14:933–944
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4, pp 9 http://palaeo-electronica.org/ 2001–1/past/issuel.0 l.htm
Horry JP, Jay M (1988) An evolutionary background of bananas as deduced from flavonoids diversification. In: Identification of genetic diversity in the genus Musa. Proceedings of an international workshop held at Los Banos, Philippines 5–10 September 1988. INIBAP, Montpellier, France, pp 41–55
Hutton MG, Loy JB (1992) Inheritance of cold germinability in muskmelon. Hort Sci 27:826–829
Jung YH, Eun YS, Seung JC et al (2004) Phylogenetic analysis of plastid trnL–trnF sequences from Arisaema species (Araceae) in Korea. Euphytica 138:81–88
Lebot V (1999) Biomolecular evidence for plant domestication in Sahul. Genet Resour Crop Evol 46:619–628
Loessl A, Goetz M, Braun A, Wenzel G (2000) Molecular markers for cytoplasm in potato: male sterility and contribution of different plastid-mitochondrial configurations to starch production. Euphytica 116:221–230
Nair AS, Teo CH, Schwarzacher T, Heslop-Harrison P (2005) Genome classification of banana cultivars from South India using IRAP markers. Euphytica 144:285–290
Nishikawa T, Salomon B, Komatsuda T, von Bothmer R, Kadowaki K (2002) Molecular phylogeny of the genus Hordeum using three chloroplast DNA sequences. Genome 45:1157–1166
Nwakanma DC, Pillay M, Okoli BE, Tenkouano A (2003) Sectional relationships in the genus Musa L. inferred from the PCR-RFLP of organelle DNA sequences. Theor Appl Gen 107:850–856
Ortiz R (2003) Analytical breeding. Acta Hort (ISHS) 622:235–247 http://www.actahort.org/books/622/622_21.htm
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Raboin ML, Carreel F, Noyer JL et al (2005) Diploid ancestors of triploid export banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Mol Breed 16:333–341
Shonnard GC, Gepts P (1994) Genetics of heat tolerance during reproductive development in common bean. Crop Sci 34:1168–1175
Simmonds NW (1995) Banana: Musa (Musaceae). In: Smartt J, Simmonds NW (eds) Evolution of crop plants, 2nd edn. Longman Scientific & Technical, Essex, pp 370–375
Simmonds NW (1962) The evolution of bananas. Longman, London
Simmonds NW, Shepherd K (1955) The taxonomy and origins of the cultivated bananas. Bot J Linn Soc 55:302–312
Sotto RC, Rabara RC (2000) Morphological diversity of Musa balbisiana Colla in the Philippines. Info Musa 9:28–30
Swangpol S, Volkaert H, Sotto RC, Seelanan T (2007) Utility of selected non-coding chloroplast sequences for lineage assessment of Musa interspecific hybrids. J Biochem Mol Biol 40:577–587
Taberlet P, Gielly L, Patou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tsumura Y, Kawahara T, Wickneswari R, Yoshimura K (1996) Molecular phylogeny of Dipterocarpaceae in Southeast Asia using RFLP of PCR amplified chloroplast genes. Theor Appl Genet 93:22–29
Ude G, Pillay M, Nwakanma D, Tenkouano A (2002) Genetic diversity in Musa acuminata Colla and Musa balbisiana Colla and some of their natural hybrids using AFLP markers. Theor Appl Genet 104:1246–1252
Voluevich EA, Buloichik A (1992) Nuclear-cytoplasmic interactions in wheat resistance to fungi pathogens: V. Quantitative resistance of alloplasmic line seedlings of Penjamo 62 variety to powdery mildew. Genetika 28:82–88
Acknowledgments
Ratri Boonruangrod and the present work were funded by Biodiversity International (formerly INIBAP). The authors are grateful to A. Burg for the excellent technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Guiderdoni.
Rights and permissions
About this article
Cite this article
Boonruangrod, R., Desai, D., Fluch, S. et al. Identification of cytoplasmic ancestor gene-pools of Musa acuminata Colla and Musa balbisiana Colla and their hybrids by chloroplast and mitochondrial haplotyping. Theor Appl Genet 118, 43–55 (2008). https://doi.org/10.1007/s00122-008-0875-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0875-3