Abstract
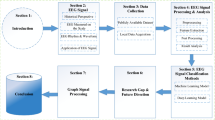
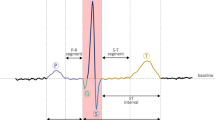
In the last 20 years, most of the human demise have been due to improper functioning of the heart. ECG is a very essential, initial clinical test for diagnosing dangerous cardiac electrical disturbances. Many techniques have been proposed to analyze and process ECG signals. It is difficult to check the performance conspicuously because the actual ECG is corrupted by several sources of noise and artifacts. In view of this, the generation of an artificial ECG signal that can reflect all the characteristics of a real ECG signal is a very challenging task in biomedical signal processing. Classification of ECG data aids early detection and prediction of any heart-related disease so that timely treatment can be suggested by the clinicians. This paper presents a method of modeling an ECG signal based on the parametric cubic splines and derives new data set therefrom, followed by ECG classification, with three different classification techniques with the aid of Orange software. The support vector machine (SVM), CN2 rule Induction and tree classifiers have been tested on the model and the Tree algorithm has shown the best performance metrics. This approach is applied for normal and abnormal sinus rhythms. Analysis and processing of these modeled ECG signals eliminate the requirement of pre-processing and conventional feature extraction techniques.




Similar content being viewed by others
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Notes
The program for developing spline model of ECG is available through the following link upon reader’s request: https://drive.google.com/file/d/10-VGZYjYvMeBoN8QEuEw5HfRvC-BrU75/view?usp=sharing
References
J. Ackora-Prah, A.Y. Aidoo, K.B. Gyamfi, An artificial ECG signal generating function in matlabtm. Appl. Math. Sci. 7(54), 2675–2686 (2013)
F.I. Alarsan, M. Younes, Analysis and classification of heart diseases using heartbeat features and machine learning algorithms. J. Big Data 6(1), 1–15 (2019)
G. Amala, Orange tool approach for comparative analysis of supervised learning algorithm in classification mining. J. Anal. Comput. 13(1), 1–10 (2019)
R. Barrio, C. Varea, J. Aragón, P. Maini, A two-dimensional numerical study of spatial pattern formation in interacting turing systems. Bull. Math. Biol. 61(3), 483–505 (1999)
J. Bernatavičienė, G. Dzemyda, O. Kurasova, D. Buteikienė, V. Barzdžiukas, A. Paunksnis, Rule induction for ophthalmological data classification. In Proceedings of EURO Mini Conference on Continuous Optimization and Knowledge-Based Technologies. Vilnius: Technika, pages 328–34 (2008)
L. Chaves, G. Marques, Data mining techniques for early diagnosis of diabetes: a comparative study. Appl. Sci. 11(5), 2218 (2021)
G.D. Clifford, A novel framework for signal representation and source separation: applications to filtering and segmentation of biosignals. J. Biol. Syst. 14(02), 169–183 (2006)
G.D. Clifford, P.E. McSharry, A realistic coupled nonlinear artificial ECG, BP, and respiratory signal generator for assessing noise performance of biomedical signal processing algorithms. Fluct. Noise Biol. Biophys. Biomed. Syst. II 5467, 290–301 (2004)
J. Demšar, T. Curk, A. Erjavec, Č Gorup, T. Hočevar, M. Milutinovič, M. Možina, M. Polajnar, M. Toplak, A. Starič et al., Orange: data mining toolbox in python. J. Mach. Learn. Res. 14(1), 2349–2353 (2013)
B. Doğan, M. Korürek, A new ECG beat clustering method based on kernelized fuzzy c-means and hybrid ant colony optimization for continuous domains. Appl. Soft Comput. 12(11), 3442–3451 (2012)
M. Engin, ECG beat classification using neuro-fuzzy network. Pattern Recogn. Lett. 25(15), 1715–1722 (2004)
M. Gidea, C. Gidea, W. Byrd, Deterministic models for simulating electrocardiographic signals. Commun. Nonlinear Sci. Numer. Simul. 16(10), 3871–3880 (2011)
A.L. Goldberger, L.A. Amaral, L. Glass, J.M. Hausdorff, P.C. Ivanov, R.G. Mark, J.E. Mietus, G.B. Moody, C.-K. Peng, H.E. Stanley, Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation 101(23), 215–220 (2000)
A. Goldberger, Z. Goldberger, A. Shvilkin, Clinical Electrocardiography: A Simplified Approach E-Book, 9th edn. (Elsevier Health Sciences, Amsterdam, 2017)
F.G. Guilak, A spline framework for optimal representation of semiperiodic signals. PhD thesis, Portland State University (2015)
N. Jafarnia-Dabanloo, D. McLernon, H. Zhang, A. Ayatollahi, V. Johari-Majd, A modified Zeeman model for producing HRV signals and its application to ECG signal generation. J. Theort. Biol. 244(2), 180–189 (2007)
S.H. Jambukia, V.K. Dabhi, H.B. Prajapati, ECG beat classification using machine learning techniques. Int. J. Biomed. Eng. Technol. 26(1), 32–53 (2018)
A. Khazaee, A. Zadeh, ECG beat classification using particle swarm optimization and support vector machine. Front. Comput. Sci. 8(2), 217–231 (2014)
M. Korürek, B. Doğan, ECG beat classification using particle swarm optimization and radial basis function neural network. Exp. Syst. Appl. 37(12), 7563–7569 (2010)
P. Kovacs, ECG signal generator based on geometrical features. Ann. Univ. Sci. Budapest Sect. Comput. 37, 247–260 (2012)
H. Li, D. Yuan, X. Ma, D. Cui, L. Cao, Genetic algorithm for the optimization of features and neural networks in ECG signals classification. Sci. Rep. 7(1), 1–12 (2017)
P.E. McSharry, G.D. Clifford, L. Tarassenko, L.A. Smith, A dynamical model for generating synthetic electrocardiogram signals. IEEE Trans. Biomed. Eng. 50(3), 289–294 (2003)
D. Noble, A modification of the Hodgkin–Huxley equations applicable to Purkinje fibre action and pacemaker potentials. J. Physiol. 160(2), 317 (1962)
M. Rozycki, T.D. Satterthwaite, N. Koutsouleris, G. Erus, J. Doshi, D.H. Wolf, Y. Fan, R.E. Gur, R.C. Gur, E.M. Meisenzahl et al., Multisite machine learning analysis provides a robust structural imaging signature of schizophrenia detectable across diverse patient populations and within individuals. Schizophr. Bull. 44(5), 1035–1044 (2018)
R. Sameni, G.D. Clifford, C. Jutten, M.B. Shamsollahi, Multichannel ECG and noise modeling: application to maternal and fetal ECG signals. EURASIP J. Adv. Signal Process. 2007, 1–14 (2007)
D.R.V. Sarangam-Kodati, Analysis of heart disease using in data mining tools orange and Weka. Global J. Comput. Sci. Technol. 18, 17–21 (2018)
I. Zeid, CAD/CAM theory and practice / Ibrahim Zeid (McGraw-Hill Series in Mechanical Engineering, New York, 1991)
F. Zhu, F. Ye, Y. Fu, Q. Liu, B. Shen, Electrocardiogram generation with a bidirectional LSTM–CNN generative adversarial network. Sci. Rep. 9(1), 1–11 (2019)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, A., Bhusnur, S. & Mishra, S. A Neoteric Parametric Representation and Classification of ECG Signal. Circuits Syst Signal Process 42, 5725–5738 (2023). https://doi.org/10.1007/s00034-023-02359-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00034-023-02359-6