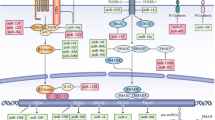
Abstract
Epithelial to mesenchymal transition (EMT) is a complex plastic and reversible cellular process that has critical roles in diverse physiological and pathological phenomena. EMT is involved in embryonic development, organogenesis and tissue repair, as well as in fibrosis, cancer metastasis and drug resistance. In recent years, the ability to edit the genome using the clustered regularly interspaced palindromic repeats (CRISPR) and associated protein (Cas) system has greatly contributed to identify or validate critical genes in pathway signaling. This review delineates the complex EMT networks and discusses recent studies that have used CRISPR/Cas technology to further advance our understanding of the EMT process.


Similar content being viewed by others
References
Brabletz T, Kalluri R, Nieto MA, Weinberg RA (2018) EMT in cancer. Nat Rev Cancer 18:128–134
Nieto MA, Huang RY, Jackson RA, Thiery JP (2016) Emt: 2016. Cell 166:21–45
Lu W, Kang Y (2019) Epithelial–mesenchymal plasticity in cancer progression and metastasis. Dev Cell 49:361–374
Chu YS et al (2006) Prototypical type I E-cadherin and type II cadherin-7 mediate very distinct adhesiveness through their extracellular domains. J Biol Chem 281:2901–2910
Pal M, Bhattacharya S, Kalyan G, Hazra S (2018) Cadherin profiling for therapeutic interventions in epithelial mesenchymal transition (EMT) and tumorigenesis. Exp Cell Res 368:137–146
Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9:265–273
Puisieux A, Brabletz T, Caramel J (2014) Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 16:488–494
Kim DH, Xing T, Yang Z, Dudek R, Lu Q, Chen Y-H (2018) Epithelial mesenchymal transition in embryonic development, tissue repair and cancer: a comprehensive overview. J Clin Med 7:1
Suhail Y, Cain MP, Vanaja K, Kurywchak PA, Levchenko A, Kalluri R (2019) Systems biology of cancer metastasis. Cell Syst 9:109–127
Shibue T, Weinberg RA (2017) EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14:611–629
Tsuji T, Ibaragi S, Hu GF (2009) Epithelial–mesenchymal transition and cell cooperativity in metastasis. Cancer Res 69:7135–7139
Labelle M, Begum S, Hynes RO (2011) Direct signaling between platelets and cancer cells induces an epithelial–mesenchymal-like transition and promotes metastasis. Cancer Cell 20:576–590
Yu M et al (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339:580–584
Aceto N et al (2014) Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158:1110–1122
Szczerba BM et al (2019) Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566:553–557
Thiery JP (2002) Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2:442–454
Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J (2012) Spatiotemporal regulation of epithelial–mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22:725–736
Huber V et al (2017) Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol 43:74–89
Peppicelli S et al (2017) The acidic microenvironment as a possible niche of dormant tumor cells. Cell Mol Life Sci 74:2761–2771
Samanta D, Semenza GL (2018) Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochim Biophys Acta Rev Cancer 1870:15–22
Schito L, Semenza GL (2016) Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer 2:758–770
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ (2008) Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol 10:295–305
Mathieu J, Zhang Z, Nelson A, Lamba DA, Reh TA, Ware C, Ruohola-Baker H (2013) Hypoxia induces re-entry of committed cells into pluripotency. Stem Cells 31:1737–1748
Azimi I, Petersen RM, Thompson EW, Roberts-Thomson SJ, Monteith GR (2017) Hypoxia-induced reactive oxygen species mediate N-cadherin and SERPINE1 expression, EGFR signalling and motility in MDA-MB-468 breast cancer cells. Sci Rep 7:15140
Azimi I, Monteith GR (2016) Plasma membrane ion channels and epithelial to mesenchymal transition in cancer cells. Endocr Relat Cancer 23:R517–R525
Azimi I (2018) The interplay between HIF-1 and calcium signalling in cancer. Int J Biochem Cell Biol 97:73–77
Andreucci E et al (2017) Carbonic anhydrase IX inhibition affects viability of cancer cells adapted to extracellular acidosis. J Mol Med 95:1341–1353
Lee SH et al (2018) Carbonic anhydrase IX is a pH-stat that sets an acidic tumour extracellular pH in vivo. Br J Cancer 119:622–630
McDonald PC, Swayampakula M, Dedhar S (2018) Coordinated regulation of metabolic transporters and migration/invasion by carbonic anhydrase IX. Metabolites 8:1–11
Ward C, Meehan J, Gray M, Kunkler IH, Langdon SP, Argyle DJ (2018) Carbonic Anhydrase IX (CAIX), cancer, and radiation responsiveness. Metabolites 8:1–18
Sedlakova O, Svastova E, Takacova M, Kopacek J, Pastorek J, Pastorekova S (2014) Carbonic anhydrase IX, a hypoxia-induced catalytic component of the pH regulating machinery in tumors. Front Physiol 4:400
Mantovani A, Allavena P, Sica A, Balkwill F (2008) Cancer-related inflammation. Nature 454:436–444
Akhurst RJ, Derynck R (2001) TGF-beta signaling in cancer–a double-edged sword. Trends Cell Biol 11:S44–51
David CJ, Massague J (2018) Contextual determinants of TGFbeta action in development, immunity and cancer. Nat Rev Mol Cell Biol 19:419–435
Derynck R, Budi EH (2019) Specificity, versatility, and control of TGF-β family signaling. Sci Signal 12:eaav5183
Heldin C-H, Vanlandewijck M, Moustakas AJFI (2012) Regulation of EMT by TGFβ in cancer. FEBS Lett 586:1959–1970
Laurenzana A et al (2015) Inhibition of uPAR-TGFbeta crosstalk blocks MSC-dependent EMT in melanoma cells. J Mol Med (Berl) 93:783–794
Guo Q (2017) Changes in mitochondrial function during EMT induced by TGFbeta-1 in pancreatic cancer. Oncol Lett 13:1575–1580
Moreno-Bueno G et al (2009) The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc 4:1591–1613
Huang RY et al (2013) An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis 4:e915
Li G, Liu Y, Yin H, Zhang X, Mo X, Tang J, Chen W (2014) E-cadherin gene promoter hypermethylation may contribute to the risk of bladder cancer among Asian populations. Gene 534:48–53
Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7:415–428
Song Y et al (2014) MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 5:11669–11680
Antony J et al (2016) The GAS6-AXL signaling network is a mesenchymal (Mes) molecular subtype-specific therapeutic target for ovarian cancer. Sci Signal 9:ra97
Aref AR et al (2013) Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integr Biol (Camb) 5:381–389
Chua KN, Sim WJ, Racine V, Lee SY, Goh BC, Thiery JP (2012) A cell-based small molecule screening method for identifying inhibitors of epithelial–mesenchymal transition in carcinoma. PLoS One 7:e33183
Chung VY et al (2016) GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep 6:19943
Jia D, Park JH, Jung KH, Levine H, Kaipparettu BA (2018) Elucidating the metabolic plasticity of cancer: mitochondrial reprogramming and hybrid metabolic states. Cells 7:21
LeBleu VS et al (2014) PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 16(992–1003):1–15
Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, Onuchic JN, Levine H (2015) Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol 5:155
Nguyen DX, Bos PD, Massague J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9:274–284
Khoo BL et al (2015) Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget 6:15578–15593
Plaks V, Koopman CD, Werb Z (2013) Circulating tumor cells. Science 341:1186–1188
Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP (2014) Epithelial–mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 6:1279–1293
Yadavalli S et al (2017) Data-driven discovery of extravasation pathway in circulating tumor cells. Sci Rep 7:43710
Fischer KR et al (2015) Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527:472–476
Zheng X et al (2015) Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527:525–530
Aiello NM, Brabletz T, Kang Y, Nieto MA, Weinberg RA, Stanger BZ (2017) Upholding a role for EMT in pancreatic cancer metastasis. Nature 547:E7–E8
Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, Yang J, Weinberg RA (2017) Upholding a role for EMT in breast cancer metastasis. Nature 547:E1–E3
Krebs AM et al (2017) The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 19:518–529
Aiello NM, Kang Y (2019) Context-dependent EMT programs in cancer metastasis. J Exp Med 216:1016–1026
Liu X, Fan D (2015) The epithelial–mesenchymal transition and cancer stem cells: functional and mechanistic links. Curr Pharm Des 21:1279–1291
Nel I, David P, Gerken GG, Schlaak JF, Hoffmann AC (2014) Role of circulating tumor cells and cancer stem cells in hepatocellular carcinoma. Hepatol Int 8:321–329
Kalluri R, Weinberg RA (2009) The basics of epithelial–mesenchymal transition. J Clin Investig 119:1420–1428
Stemmler MP, Eccles RL, Brabletz S, Brabletz T (2019) Non-redundant functions of EMT transcription factors. Nat Cell Biol 21:102–112
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196
Yang J, Weinberg RA (2008) Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14:818–829
Pei D, Shu X, Gassama-Diagne A, Thiery JP (2019) Mesenchymal–epithelial transition in development and reprogramming. Nat Cell Biol 21:44
Dongre A, Weinberg RA (2018) New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20:69–84
Zhang Y, Weinberg RA (2018) Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med 12:361–373
Hay ED (2005) The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn 233:706–720
Timmerman LA et al (2004) Notch promotes epithelial–mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 18:99–115
Leask A, Abraham DJ (2004) TGF-beta signaling and the fibrotic response. FASEB J 18:816–827
Kaimori A, Potter J, Kaimori JY, Wang C, Mezey E, Koteish A (2007) Transforming growth factor-beta1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro. J Biol Chem 282:22089–22101
Saika S et al (2009) TGF beta in fibroproliferative diseases in the eye. Front Biosci (Schol Ed) 1:376–390
Kriz W, Kaissling B, Le Hir M (2011) Epithelial–mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J Clin Investig 121:468–474
Lovisa S et al (2015) Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med 21:998–1009
Grande MT et al (2015) Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med 21:989–997
Bi WR, Yang CQ, Shi Q (2012) Transforming growth factor-beta1 induced epithelial–mesenchymal transition in hepatic fibrosis. Hepatogastroenterology 59:1960–1963
Pourgholamhossein F et al (2018) Pirfenidone protects against paraquat-induced lung injury and fibrosis in mice by modulation of inflammation, oxidative stress, and gene expression. Food Chem Toxicol 112:39–46
Mamuya FA, Wang Y, Roop VH, Scheiblin DA, Zajac JC, Duncan MK (2014) The roles of alphaV integrins in lens EMT and posterior capsular opacification. J Cell Mol Med 18:656–670
Chang KC, Petrash JM (2015) Aldose reductase mediates transforming growth factor beta2 (TGF-beta2)-induced migration and epithelial-to-mesenchymal transition of lens-derived epithelial cells. Investig Ophthalmol Vis Sci 56:4198–4210
Chang KC, Shieh B, Petrash JM (2017) Influence of aldose reductase on epithelial-to-mesenchymal transition signaling in lens epithelial cells. Chem Biol Interact 276:149–154
Nahomi RB, Pantcheva MB, Nagaraj RH (2016) alphaB-crystallin is essential for the TGF-beta2-mediated epithelial to mesenchymal transition of lens epithelial cells. Biochem J 473:1455–1469
Teven CM, Farina EM, Rivas J, Reid RR (2014) Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes Dis 1:199–213
Correia AC, Moonen JR, Brinker MG, Krenning G (2016) FGF2 inhibits endothelial-mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-beta signaling. J Cell Sci 129:569–579
Ciruna B, Rossant J (2001) FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell 1:37–49
Pera EM, Ikeda A, Eivers E, De Robertis EM (2003) Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev 17:3023–3028
Wu C et al (2016) Interaction between Wnt/beta-catenin pathway and microRNAs regulates epithelial–mesenchymal transition in gastric cancer (Review). Int J Oncol 48:2236–2246
Du B, Shim JS (2016) Targeting epithelial–mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules 21:1–15
Roy M, Pear WS, Aster JC (2007) The multifaceted role of Notch in cancer. Curr Opin Genet Dev 17:52–59
Wels C, Joshi S, Koefinger P, Bergler H, Schaider H (2011) Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial–mesenchymal transition-like phenotype in melanoma. J Investig Dermatol 131:1877–1885
Sinh ND, Endo K, Miyazawa K, Saitoh M (2017) Ets1 and ESE1 reciprocally regulate expression of ZEB1/ZEB2, dependent on ERK1/2 activity, in breast cancer cells. Cancer Sci 108:952–960
Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP (2004) Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J 23:1155–1165
Chang K-C et al (2019) Opposing effects of growth and differentiation factors in cell-fate specification. Curr Biol 29:1963–1975
Yuan X et al (2014) Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J Hematol Oncol 7:87
Tan C, Hu W, He Y, Zhang Y, Zhang G, Xu Y, Tang J (2018) Cytokine-mediated therapeutic resistance in breast cancer. Cytokine 108:151–159
Wang J, Knaut H (2014) Chemokine signaling in development and disease. Development 141:4199–4205
Maritzen T, Schachtner H, Legler DF (2015) On the move: endocytic trafficking in cell migration. Cell Mol Life Sci 72:2119–2134
Rokavec M et al (2014) IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Investig 124:1853–1867
Antoon JW et al (2012) Altered death receptor signaling promotes epithelial-to-mesenchymal transition and acquired chemoresistance. Sci Rep 2:539
Sonego M et al (2019) USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci Adv 5:eaav3235
van Staalduinen J, Baker D, ten Dijke P, van Dam H (2018) Epithelial–mesenchymal-transition-inducing transcription factors: new targets for tackling chemoresistance in cancer? Oncogene 37:6195–6211
Peebles CL, Perlman P, Mecklenburg K, Petrillo M, Tabor J, Jarrell K, Cheng H-L (1986) A self-splicing RNA excises an intron lariat. Cell 44:213–223
Carninci P et al (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563
Park S-M, Gaur AB, Lengyel E, Peter ME (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22:894–907
Moreno-Bueno G, Portillo F, Cano A (2008) Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 27:6958–6969
Arun K, Arunkumar G, Bennet D, Chandramohan SM, Murugan AK, Munirajan AK (2018) Comprehensive analysis of aberrantly expressed lncRNAs and construction of ceRNA network in gastric cancer. Oncotarget 9:18386
Tamagawa S, Beder LB, Hotomi M, Gunduz M, Yata K, Grenman R, Yamanaka N (2014) Role of miR-200c/miR-141 in the regulation of epithelial–mesenchymal transition and migration in head and neck squamous cell carcinoma. Int J Mol Med 33:879–886
Chen H-Y et al (2012) miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Can Res 72:3631–3641
Martello G et al (2010) A MicroRNA targeting dicer for metastasis control. Cell 141:1195–1207
Zhang Y et al (2015) miR-103/107 modulates multidrug resistance in human gastric carcinoma by downregulating Cav-1. Tumor Biol 36:2277–2285
Bornachea O et al (2012) EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Sci Rep 2:434
Chang C-J et al (2011) Let-7d functions as novel regulator of epithelial–mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep 26:1003–1010
Manikandan M, Rao AKDM, Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R, Munirajan AK (2016) Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer 15:28
Manikandan M, Rao DM, Arunagiri K, Rajkumar KS, Rajaraman R, Munirajan AK (2015) Altered levels of miR-21, miR-125b-2*, miR-138, miR-155, miR-184, and miR-205 in oral squamous cell carcinoma and association with clinicopathological characteristics. J Oral Pathol Med 44:792–800
Lin C-H, Chiang M-C, Chen Y-J (2018) Microrna-328 inhibits migration and epithelial–mesenchymal transition by targeting cD44 in nasopharyngeal carcinoma cells. Oncotargets Ther 11:2375
Luo H, Liang C (2018) MicroRNA-148b inhibits proliferation and the epithelial–mesenchymal transition and increases radiosensitivity in non-small cell lung carcinomas by regulating ROCK1. Exp Ther Med 15:3609–3616
Wang J, Wang X, Liu F, Fu Y (2017) microRNA-335 inhibits colorectal cancer HCT116 cells growth and epithelial–mesenchymal transition (EMT) process by targeting Twist1. Die Pharmazie Int J Pharm Sci 72:475–481
Liu H, Pan Y, Han X, Liu J, Li R (2017) Microrna-216a promotes the metastasis and epithelial–mesenchymal transition of ovarian cancer by suppressing the PTen/aKT pathway. Oncotargets Ther 10:2701
Lu C, Shan Z, Hong J, Yang L (2017) MicroRNA-92a promotes epithelial–mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis. Int J Oncol 51:235–244
Zhang N, Tian L, Miao Z, Guo N (2018) MicroRNA-197 induces epithelial–mesenchymal transition and invasion through the downregulation of HIPK2 in lung adenocarcinoma. J Genet 97:47–53
Song X-F, Wang Q-H, Huo R (2017) Effects of microRNA-708 on epithelial–mesenchymal transition, cell proliferation and apoptosis in melanoma cells by targeting LEF1 through the Wnt signaling pathway. Pathol Oncol Res 25:377–389
Kung JT, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193:651–669
Arunkumar G, Murugan AK, Rao PSH, Subbiah S, Rajaraman R, Munirajan AK (2017) Long non-coding RNA CCAT1 is overexpressed in oral squamous cell carcinomas and predicts poor prognosis. Biomed Rep 6:455–462
Xu Y, Yao Y, Qin W, Zhong X, Jiang X, Cui Y (2018) Long non-coding RNA CCAT2 promotes cholangiocarcinoma cells migration and invasion by induction of epithelial-to-mesenchymal transition. Biomed Pharmacother 99:121–127
Kawasaki N, Miwa T, Hokari S, Sakurai T, Ohmori K, Miyauchi K, Miyazono K, Koinuma D (2018) Long noncoding RNA NORAD regulates transforming growth factor-β signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci 109:2211–2220
Du D, Lian D, Amin B, Yan W (2017) Long non-coding RNA CRNDE is a novel tumor promoter by modulating PI3K/AKT signal pathways in human gastric cancer. Eur Rev Med Pharmacol Sci 21:5392–5398
Lv W-Q, Wang H-C, Peng J, Wang Y-X, Jiang J-H, Li C-Y (2017) Gene editing of the extra domain A positive fibronectin in various tumors, amplified the effects of CRISPR/Cas system on the inhibition of tumor progression. Oncotarget 8:105020
Zarringhalam K, Tay Y, Kulkarni P, Bester AC, Pandolfi PP, Kulkarni RV (2017) Identification of competing endogenous RNAs of the tumor suppressor gene PTEN: a probabilistic approach. Sci Rep 7:7755
Li C et al (2018) Long non-coding RNA XIST promotes TGF-β-induced epithelial–mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett 418:185–195
Dasgupta P et al (2018) MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther 17:1061–1069
Ge Y et al (2015) fMiRNA-192 and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet 11:e1005726
Shapiro RS, Chavez A, Collins JJ (2018) CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat Rev Microbiol 16:333–339
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Sander JD, Joung JK (2014) CRISPR–Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347
Cho SW, Kim S, Kim JM, Kim J-S (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230
Cong L et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. eLife 2:1–9
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
La Russa MF, Qi LS (2015) The new state of the art: Cas9 for gene activation and repression. Mol Cell Biol 35:3800–3809
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Gilbert LA et al (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661
Hilton IB, D'ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510
Klann TS et al (2017) CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol 35:561
Liu XS et al (2016) Editing DNA methylation in the mammalian genome. Cell 167(233–247):e17
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 551:464
Cox DB, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358:1019–1027
Adli M (2018) The CRISPR tool kit for genome editing and beyond. Nat Commun 9:1911
Sánchez-Rivera FJ, Jacks T (2015) Applications of the CRISPR–Cas9 system in cancer biology. Nat Rev Cancer 15:387
Zhang L et al (2020) FRK plays an oncogenic role in non-small cell lung cancer by enhancing the stemness phenotype via induction of metabolic reprogramming. Int J Cancer 146:208–222
Xu X et al (2017) CRISPR-ON-Mediated KLF4 overexpression inhibits the proliferation, migration and invasion of urothelial bladder cancer in vitro and in vivo. Oncotarget 8:102078–102087
Shibata T, Shibata S, Ishigaki Y, Kiyokawa E, Ikawa M, Singh DP, Sasaki H, Kubo E (2018) Tropomyosin 2 heterozygous knockout in mice using CRISPR-Cas9 system displays the inhibition of injury-induced epithelial–mesenchymal transition, and lens opacity. Mech Ageing Dev 171:24–30
Haraguchi M, Sato M, Ozawa M (2015) CRISPR/Cas9n-mediated deletion of the Snail 1Gene (SNAI1) reveals its role in regulating cell morphology, cell–cell interactions, and gene expression in ovarian cancer (RMG-1) cells. PLoS One 10:e0132260
Wang HC, Yang Y, Xu SY, Peng J, Jiang JH, Li CY (2015) The CRISPR/Cas system inhibited the pro-oncogenic effects of alternatively spliced fibronectin extra domain A via editing the genome in salivary adenoid cystic carcinoma cells. Oral Dis 21:608–618
Zhao G et al (2017) Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget 8:94666
McFaline-Figueroa JL, Hill AJ, Qiu X, Jackson D, Shendure J, Trapnell C (2019) A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat Genet 51:1389–1398
Qu X et al (2019) c-Myb promotes growth and metastasis of colorectal cancer through c-fos-induced epithelial–mesenchymal transition. Cancer Sci 110:3183–3196
Freihen V et al (2019) SNAIL1 employs β-Catenin-LEF1 complexes to control colorectal cancer cell invasion and proliferation. Int J Cancer. https://doi.org/10.1002/ijc.32644
Zeng K et al (2018) LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2-mediated p53 ubiquitination and degradation. Oncogene 37:5534–5551
Liu Z et al (2019) Tn antigen promotes human colorectal cancer metastasis via H-Ras mediated epithelial–mesenchymal transition activation. J Cell Mol Med 23:2083–2092
Jubair L, Fallaha S, McMillan NA (2019) Systemic delivery of CRISPR/Cas9 targeting HPV oncogenes is effective at eliminating established tumors. Mol Ther 27:2091–2099
Wilbie D, Walther J, Mastrobattista E (2019) Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc Chem Res 52:1555–1564
Tong S, Moyo B, Lee CM, Leong K, Bao G (2019) Engineered materials for in vivo delivery of genome-editing machinery. Nat Rev Mater 4:726–737
Liu Q et al (2019) Multistage delivery nanoparticle facilitates efficient CRISPR/dCas9 activation and tumor growth suppression in vivo. Adv Sci 6:1–12
Luther D, Lee Y, Nagaraj H, Scaletti F, Rotello V (2018) Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin Drug Deliv 15:905–913
Biagioni A, Laurenzana A, Margheri F, Chillà A, Fibbi G (2018) Delivery systems of CRISPR/Cas9-based cancer gene therapy. J Biol Eng 12:33
Lu Y, Liang M, Zhang Q, Liu Z, Song Y, Lai L, Li Z (2019) Mutations of GADD45G in rabbits cause cleft lip by the disorder of proliferation, apoptosis and epithelial–mesenchymal transition (EMT). Biochimica et Biophysica Acta (BBA) Mol Basis Dis 1865:2356–2367
Seok H, Lee H, Jang E-S, Chi SW (2018) Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol Life Sci 75:797–814
Spagnou S, Miller AD, Keller M (2004) Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry 43:13348–13356
Zhou J, Shum K-T, Burnett JC, Rossi JJ (2013) Nanoparticle-based delivery of RNAi therapeutics: progress and challenges. Pharmaceuticals 6:85–107
Fedorov Y, King A, Anderson E, Karpilow J, Ilsley D, Marshall W, Khvorova A (2005) Different delivery methods—different expression profiles. Nat Methods 2:241
Luo J (2016) CRISPR/Cas9: from genome engineering to cancer drug discovery. Trends Cancer 2:313–324
Goyal A, Myacheva K, Groß M, Klingenberg M, Duran Arqué B, Diederichs SJN (2017) Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res 45:e12–e12
Chen F, Ji J, Shen J, Lu XJ (2017) When long noncoding RNAs meet genome editing in pluripotent stem cells. Stem Cells Int 2017:1–13
Szafranski P, Karolak JA, Lanza D, Gajęcka M, Heaney J, Stankiewicz P (2017) CRISPR/Cas9-mediated deletion of lncRNA Gm26878 in the distant Foxf1 enhancer region. Mamm Genome 28:275–282
Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K (2014) Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32:267–273
Zare K, Shademan M, Seno MMG, Dehghani H (2018) CRISPR/Cas9 knockout strategies to ablate CCAT1 lncRNA gene in cancer cells. Biol Proced Online 20:21
Jiang Q, Meng X, Meng L, Chang N, Xiong J, Cao H, Liang Z (2014) Small indels induced by CRISPR/Cas9 in the 5′ region of microRNA lead to its depletion and Drosha processing retardance. RNA Biol 11:1243–1249
Zhao Y, Dai Z, Liang Y, Yin M, Ma K, He M, Ouyang H, Teng C-B (2014) Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci Rep 4:3943
Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y (2016) CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 6:22312
Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM (2009) Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res 37:2584–2595
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133:647–658
Sabatel C et al (2011) MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One 6:e16979
Kim YJ, Hwang SJ, Bae YC, Jung JS (2009) MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem cells 27:3093–3102
Papagiannakopoulos T, Shapiro A, Kosik KS (2008) MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Can Res 68:8164–8172
Huo W et al (2017) Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer 8:57
Yoshino H, Yonemori M, Miyamoto K, Tatarano S, Kofuji S, Nohata N, Nakagawa M, Enokida H (2017) microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 8:20881
Hoffmann MD et al (2019) Cell-specific CRISPR–Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res 47:e75–e75
Voets O et al (2017) Highly efficient gene inactivation by adenoviral CRISPR/Cas9 in human primary cells. PLoS One 12:e0182974
Fang S et al (2018) circHECTD1 promotes the silica-induced pulmonary endothelial–mesenchymal transition via HECTD1. Cell Death Dis 9:396
Jiang R et al (2019) The emerging roles of a novel CCCH-type zinc finger protein, ZC3H4, in silica-induced epithelial to mesenchymal transition. Toxicol Lett 307:26–40
Tedja R et al (2019) Protein kinase Cα–mediated phosphorylation of Twist1 at Ser-144 prevents Twist1 ubiquitination and stabilizes it. J Biol Chem 294:5082–5093
Isaji T, Im S, Kameyama A, Wang Y, Fukuda T, Gu J (2019) A complex between phosphatidylinositol 4-kinase IIα and integrin α3β1 is required for N-glycan sialylation in cancer cells. J Biol Chem 294:4425–4436
Wang X, Zhang W, Ding Y, Guo X, Yuan Y, Li D (2017) CRISPR/Cas9-mediated genome engineering of CXCR4 decreases the malignancy of hepatocellular carcinoma cells in vitro and in vivo. Oncol Rep 37:3565–3571
He J, Zhang W, Li A, Chen F, Luo RJB (2018) Knockout of NCOA5 impairs proliferation and migration of hepatocellular carcinoma cells by suppressing epithelial-to-mesenchymal transition. Biochem Biophys Res Commun 500:177–183
Qi X-K et al (2018) OVOL2 links stemness and metastasis via fine-tuning epithelial–mesenchymal transition in nasopharyngeal carcinoma. Theranostics 8:2202–2216
Jägle S, Dertmann A, Schrempp M, Hecht A (2017) ZEB1 is neither sufficient nor required for epithelial–mesenchymal transition in LS174T colorectal cancer cells. Biochem Biophys Res Commun 482:1226–1232
Dong X, Jiang Y, Liu J, Liu Z, Gao T, An G, Wen T (2018) T-Synthase deficiency enhances oncogenic features in human colorectal cancer cells via activation of epithelial–mesenchymal transition. Biomed Res Int 2018:1–7
Franke FC et al (2019) The tumor suppressor SASH1 interacts with the signal adaptor CRKL to inhibit epithelial–mesenchymal transition and metastasis in colorectal cancer. Cell Mol Gastroenterol Hepatol 7:33–53
Wang X et al (2018) The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol Cancer 17:110
Zhou M, Liu X, Li Z, Huang Q, Li F, Li CY (2018) Caspase-3 regulates the migration, invasion and metastasis of colon cancer cells. Int J Cancer 143:921–930
Hosain SB et al (2016) Inhibition of glucosylceramide synthase eliminates the oncogenic function of p53 R273H mutant in the epithelial–mesenchymal transition and induced pluripotency of colon cancer cells. Oncotarget 7:60575
Schokrpur S et al (2016) CRISPR-mediated VHL knockout generates an improved model for metastatic renal cell carcinoma. Sci Rep 6:29032
Lin Y et al (2015) PIK3R1 negatively regulates the epithelial–mesenchymal transition and stem-like phenotype of renal cancer cells through the AKT/GSK3β/CTNNB1 signaling pathway. Sci Rep 5:8997
Belvedere R et al (2018) miR-196a is able to restore the aggressive phenotype of annexin A1 knock-out in pancreatic cancer cells by CRISPR/Cas9 genome editing. Int J Mol Sci 19:1–15
Pessolano E et al (2018) Annexin A1 may induce pancreatic cancer progression as a key player of extracellular vesicles effects as evidenced in the in vitro MIA PaCa-2 model system. Int J Mol Sci 19:3878
Santoro R, Zanotto M, Carbone C, Piro G, Tortora G, Melisi D (2018) MEKK3 sustains EMT and stemness in pancreatic cancer by regulating YAP and TAZ transcriptional activity. Anticancer Res 38:1937–1946
Zhang P et al (2018) Knockdown of survivin results in inhibition of epithelial to mesenchymal transition in retinal pigment epithelial cells by attenuating the TGFβ pathway. Biochem Biophys Res Commun 498:573–578
Liu B et al (2019) Blockade of MDM2 with inactive Cas9 prevents epithelial to mesenchymal transition in retinal pigment epithelial cells. Lab Investig 99:1874–1886
Tordjman J, Majumder M, Amiri M, Hasan A, Hess D, Lala PK (2019) Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer 19:561
Fröse J, Chen MB, Hebron KE, Reinhardt F, Hajal C, Zijlstra A, Kamm RD, Weinberg RA (2018) Epithelial–mesenchymal transition induces podocalyxin to promote extravasation via ezrin signaling. Cell Rep 24:962–972
Wu C-L et al (2018) BECN1-knockout impairs tumor growth, migration and invasion by suppressing the cell cycle and partially suppressing the epithelial–mesenchymal transition of human triple-negative breast cancer cells. Int J Oncol 53:1301–1312
Hodge DQ, Cui J, Gamble MJ, Guo WJ (2018) Histone variant macroH2A1 plays an isoform-specific role in suppressing epithelial–mesenchymal transition. Sci Rep 8:841
Røsland GV et al (2019) Epithelial to mesenchymal transition (EMT) is associated with attenuation of succinate dehydrogenase (SDH) in breast cancer through reduced expression of SDHC. Cancer Metab 7:6
Riaz SK, Ke Y, Wang F, Kayani MA, Malik MFA (2019) Influence of SHH/GLI1 axis on EMT mediated migration and invasion of breast cancer cells. Sci Rep 9:6620
Sun Y, Gao X, Wu P, Wink M, Li J, Dian L, Liang Z (2019) Jatrorrhizine inhibits mammary carcinoma cells by targeting TNIK mediated Wnt/β-catenin signalling and epithelial–mesenchymal transition (EMT). Phytomedicine 63:153015
Ramachandran A et al (2018) TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife 7:e31756
Sossey-Alaoui K, Pluskota E, Szpak D, Schiemann WP, Plow EF (2018) The Kindlin-2 regulation of epithelial-to-mesenchymal transition in breast cancer metastasis is mediated through miR-200b. Sci Rep 8:7360
Maturi V, Enroth S, Heldin CH, Moustakas A (2018) Genome-wide binding of transcription factor ZEB1 in triple-negative breast cancer cells. J Cell Physiol 233:7113–7127
Maturi V, Morén A, Enroth S, Heldin CH, Moustakas A (2018) Genomewide binding of transcription factor Snail1 in triple-negative breast cancer cells. Mol Oncol 12:1153–1174
Kumar S, Davra V, Obr AE, Geng K, Wood TL, De Lorenzo MS, Birge RB (2018) Crk adaptor protein promotes PD-L1 expression, EMT and immune evasion in a murine model of triple-negative breast cancer. Oncoimmunology 7:e1376155
Beck C et al (2019) PARP3, a new therapeutic target to alter Rictor/mTORC2 signaling and tumor progression in BRCA1-associated cancers. Cell Death Differ 26:1615–1630
Kobayashi W, Ozawa M (2018) The epithelial–mesenchymal transition induced by transcription factor LEF-1 is independent of β-catenin. Biochem Biophys Rep 15:13–18
Ma Y et al (2018) CD146 mediates an E-cadherin-to-N-cadherin switch during TGF-β signaling-induced epithelial–mesenchymal transition. Cancer Lett 430:201–214
Jacob F et al (2018) Transition of mesenchymal and epithelial cancer cells depends on α1-4 galactosyltransferase-mediated glycosphingolipids. Cancer Res 2223:2017
Haraguchi M, Fukushige T, Kanekura T, Ozawa MJB (2019) E-cadherin loss in RMG-1 cells inhibits cell migration and its regulation by Rho GTPases. Biochem Biophys Rep 18:100650
Ji L et al (2018) Knockout of MTF1 inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer 9:4578–4585
Jiang X et al (2018) Metastatic prostate cancer-associated P62 inhibits autophagy flux and promotes epithelial to mesenchymal transition by sustaining the level of HDAC6. Prostate 78:426–434
Huang G et al (2016) TGF-β signal rewiring sustains epithelial–mesenchymal transition of circulating tumor cells in prostate cancer xenograft hosts. Oncotarget 7:77124
Katikireddy KR et al (2018) NQO1 downregulation potentiates menadione-induced endothelial-mesenchymal transition during rosette formation in Fuchs endothelial corneal dystrophy. Free Radic Biol Med 116:19–30
Sommerova L, Ondrouskova E, Vojtesek B, Hrstka R (2017) Suppression of AGR2 in a TGF-β-induced Smad regulatory pathway mediates epithelial–mesenchymal transition. BMC Cancer 17:546
Chockley PJ, Chen J, Chen G, Beer DG, Standiford TJ, Keshamouni VG (2018) Epithelial–mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer. J Clin Investig 128:1384–1396
Liu Y et al (2019) Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat Commun 10:1637
Vad-Nielsen J, Gammelgaard KR, Daugaard TF, Nielsen AL (2019) Cause-and-effect relationship between FGFR1 expression and epithelial–mesenchymal transition in EGFR-mutated non-small cell lung cancer cells. Lung Cancer 132:132–140
Perumal E, Youn KS, Sun S, Seung-Hyun J, Suji M, Jieying L, Yeun-Jun C (2019) PTEN inactivation induces epithelial–mesenchymal transition and metastasis by intranuclear translocation of β-catenin and snail/slug in non-small cell lung carcinoma cells. Lung Cancer 130:25–34
Raoof S et al (2019) Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene 38:6399–6413
Zhang Y-Q, Pei J-H, Shi S-S, Guo X-S, Cui G-Y, Li Y-F, Zhang H-P, Hu W-Q (2019) CRISPR/Cas9-mediated knockout of the PDEF gene inhibits migration and invasion of human gastric cancer AGS cells. Biomed Pharmacother 111:76–85
Paulitschke V et al (2019) Proteomic identification of a marker signature for MAPKi resistance in melanoma. EMBO J 38:e95874
Deiana M et al (2018) New insights into the runt domain of RUNX2 in melanoma cell proliferation and migration. Cells 7:220
Cecconi D et al (2019) Runx2 stimulates neoangiogenesis through the Runt domain in melanoma. Sci Rep 9:8052
Kim HS et al (2017) Endothelial-derived interleukin-6 induces cancer stem cell motility by generating a chemotactic gradient towards blood vessels. Oncotarget 8:100339–100352
Sui A, Xu Y, Yang J, Pan B, Wu J, Guo T, Shen Y, Guo X (2019) The histone H3 Lys 27 demethylase KDM6B promotes migration and invasion of glioma cells partly by regulating the expression of SNAI1. Neurochem Int 124:123–129
Tome-Garcia J et al (2018) Analysis of chromatin accessibility uncovers TEAD1 as a regulator of migration in human glioblastoma. Nat Commun 9:4020
Zhang Q et al (2019) Acidic bile salts induce epithelial to mesenchymal transition via VEGF signaling in non-neoplastic Barrett’s cells. Gastroenterology 156(130–144):e10
Abudureheman A et al (2018) High MLL2 expression predicts poor prognosis and promotes tumor progression by inducing EMT in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol 144:1025–1035
Frausto RF et al (2019) ZEB1 insufficiency causes corneal endothelial cell state transition and altered cellular processing. PLoS One 14:e0218279
Liu Y, Dakou E, Meng Y, Leyns L (2019) Loss of Emp2 compromises cardiogenic differentiation in mouse embryonic stem cells. Biochem Biophys Res Commun 511:173–178
Eskildsen TV et al (2019) MESP1 knock-down in human iPSC attenuates early vascular progenitor cell differentiation after completed primitive streak specification. Dev Biol 445:1–7
Soderquist RS et al (2018) Systematic mapping of BCL-2 gene dependencies in cancer reveals molecular determinants of BH3 mimetic sensitivity. Nat Commun 9:3513
Hu Q et al (2019) LncRNAs-directed PTEN enzymatic switch governs epithelial–mesenchymal transition. Cell Res 29:286–304
Santamaria PG, Moreno-Bueno G, Portillo F, Cano A (2017) EMT: present and future in clinical oncology. Mol Oncol 11:718–738
Acknowledgements
The authors would like to thank Dr. Yamini Dalal (National Cancer Institute) for constructive criticism of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mohammadinejad, R., Biagioni, A., Arunkumar, G. et al. EMT signaling: potential contribution of CRISPR/Cas gene editing. Cell. Mol. Life Sci. 77, 2701–2722 (2020). https://doi.org/10.1007/s00018-020-03449-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03449-3