Abstract
The interferon alpha-responsive gene (Ifrg15) mRNA is highly expressed in various stages during preimplantation mammalian embryo development. Unfortunately, few studies have investigated the effect of Ifrg15 in this process. In mammals, the fusion of male and female pronuclei generates a diploid zygote, and is an important step for subsequent cleavage and blastocyst formation. Here, by using RNA interference, rescue experiments, immunofluorescence staining and live cell observations, we found that preimplantation embryo development was arrested at the 1-cell stage after knocking down Ifrg15 expression. This induced DNA damage and prevented the cleavage of embryos. Furthermore, the effect of Ifrg15 deficiency in arresting preimplantation embryo development produced by specific short interfering RNA microinjection was concentration-dependent. Using transcriptome expression profiles, gene ontogeny functional annotation and enrichment analysis, we gained 197 enriched pathways based on 1445 differentially expressed genes (DEGs). Of these, 12 pathways and about one third of the DEGs were involved in DNA damage, DNA repair, cell cycle, and developmental processes. Thus, the IFRG15 protein might be an important molecule for maintaining genomic integrity and stability through upregulating or downregulating a cascade of genes to permit normal preimplantation embryo development.








Similar content being viewed by others
References
Cedars M (2005) Introduction to infertility. In: Cedars M (ed) Infertility. McGraw-Hill, New York
Plachot M (2000) Fertilization. Hum Reprod 15(Suppl 4):19–30
Wright G et al (1990) Observations on the morphology of pronuclei and nucleoli in human zygotes and implications for cryopreservation. Hum Reprod 5(1):109–115
Johnson M (2007) Essential reproduction, 6th edn. Blackwell, Oxford
Wilson EB (1925) The cell in development and heredity. Macmillan, New York
Mayer W et al (2000) Spatial separation of parental genomes in preimplantation mouse embryos. J Cell Biol 148(4):629–634
Wang S et al (2010) Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA 107(41):17639–17644
Yamaza H et al (2001) Detection of differentially expressed genes in the early developmental stage of the mouse mandible. Int J Dev Biol 45(4):675–680
Wu FR et al (2012) Sequence analysis, expression patterns and transcriptional regulation of mouse Ifrg15 during preimplantation embryonic development. Gene 507(2):119–124
Qi B et al (2007) Cloning and analysis of IFRG (interferon responsive gene) in rabbit oocytes and preimplantation embryos. Biocell 31(2):199–203
Zeng F, Schultz RM (2005) RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 283(1):40–57
Kigami D et al (2003) MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 68(2):651–654
Li L, Zheng P, Dean J (2010) Maternal control of early mouse development. Development 137(6):859–870
Kuo LJ, Yang LX (2008) Gamma-H2AX—a novel biomarker for DNA double-strand breaks. Vivo 22(3):305–309
Xiao M et al (2013) Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol 166(3):122–134
Schefe JH et al (2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J Mol Med (Berl) 84(11):901–910
Acknowledgments
The authors thank all the members of the team for giving technical support and valuable suggestions. We gratefully acknowledge Liwen Bianji for editing the article. This research was supported by NSFC (Grant Nos.: 81270701, 81401204, 81471457).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Western blot analysis showed that the anti-IFRG15 antibody was not specific (JPEG 219 kb)
Figure S2
(A) The LULL protein has no homologous sequences with IFRG15. (B) Lull1 mRNA was expressed in the GV, MII, zygote, 2-cell, 4-cell, 8-cell, morula and blastocyst stages. Lull1 showed the lowest expression in MII and zygote stages, peaked at the 2-cell stage then reduced gradually. Three batches of data were analyzed statistically. (C) Verifying the efficiency of Lull1 knockdown by qRT-PCR. There was no statistically significant difference between normal and control embryos injected with nonsilencing siRNA. Compared with the normal and control groups, sharp decreases in the expression levels of Lull1mRNA were found in those injected with Lull1 siRNA #1 (21%,p<0.05) or siRNA #2 (19%,p<0.05). (D) Comparison of embryo formation rate between control and Lull1 knockdown groups. There were no statistically significant differences (JPEG 905 kb)
Figure S3
Various concentrations of Ifrg15-specific siRNAs were microinjected: 20µM, 50 µM and 100µM. Almost all embryos in the normal and negative control group (100µM of a non-silence siRNA fragment) developed to the 2-cell stage (99% and 100%, respectively). Those microinjected with 20µM(siRNA1#, 97%, p<0.05; siRNA2#, 98%, p<0.05), 50µM(siRNA1#, 90%, p<0.05; siRNA2#, 85%, p<0.01) and 100µM(siRNA1#, 47%, p<0.001; siRNA2#, 43%, p<0.001) of siRNA specific against Ifrg15 partly developed into 2-cell embryos, respectively, and the blastocyst formation rate decreased respectively, 20µM(siRNA1#, 42%, p<0.01; siRNA2#, 32%, p<0.001); 50µM(siRNA1#, 33%, p<0.001; siRNA2#, 25%, p<0.001); 100µM(siRNA1#, 4%, p<0.001; siRNA2#, 2%, p<0.001); versus negative control (JPEG 304 kb)
Figure S4
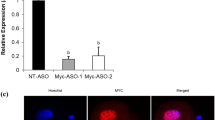
After microinjection of Ifrg15 mRNA, the expression levels of Ifrg15 were detected by immunofluorescence staining and confocal microscopy. Staining for Myc showed that positive signals were mainly located in the cytoplasm when Ifrg15 mRNA was injected. The signals for IFRG15 showed a higher expression level (JPEG 691 kb)
Figure S5
Representative genes selected from upregulated or downregulated DEGs for subsequent qRT-PCR experiments (JPEG 1712 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Wang, J., Zhao, C. et al. The interferon α-responsive gene, Ifrg15, plays vital roles during mouse early embryonic development. Cell. Mol. Life Sci. 73, 2969–2984 (2016). https://doi.org/10.1007/s00018-016-2150-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2150-0