Abstract
Stem cells provide fascinating prospects for biomedical applications by combining the ability to renew themselves and to differentiate into specialized cell types. Since the first isolation of embryonic stem (ES) cells about 30 years ago, there has been a series of groundbreaking discoveries that have the potential to revolutionize modern life science. For a long time, embryos or germ cell-derived cells were thought to be the only source of pluripotency—a dogma that has been challenged during the last decade. Several findings revealed that cell differentiation from (stem) cells to mature cells is not in fact an irreversible process. The molecular mechanism underlying cellular reprogramming is poorly understood thus far. Identifying how pluripotency maintenance takes place in ES cells can help us to understand how pluripotency induction is regulated. Here, we review recent advances in the field of stem cell regulation focusing on key transcription factors and their functional interplay with non-coding RNAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the first isolation of embryonic stem cells about 30 years ago, there has been a series of fundamental developments in stem cell biology that have the potential to revolutionize modern biomedicine. Stem cells exhibit very special cellular properties by combining the ability to renew themselves and to differentiate into specialized cell types. Adult or somatic stem cells reside in multiple organs of the organism, such as bone marrow, skin and brain (for review, see [1, 2]). They can mature into a restricted subset (or even just a single type) of specialized cells—a cellular property designated as multi- and unipotency, respectively (see Table 1 for definitions). Whereas somatic stem cells exhibit a restricted differentiation potential, Evans and Kaufman succeeded in establishing a very particular stem cell type from an embryonic source [3] that is able to differentiate into any cell type of the organism (pluripotency). These so-called embryonic stem (ES) cells originate from the inner cell mass (ICM) of blastocysts. Totipotent cells, in contrast, can contribute to both embryonic and extraembryonic tissue formation (Fig. 1). For a long time, embryos or germ cell-derived cells were thought to be the only source of pluripotency—a dogma that has been challenged during the last decade. About 10 years ago, Wilmut et al. reported the first production of a normal adult mammalian (the sheep, ‘Dolly’) by transplanting the nuclei of ex vivo cultured mammary gland cells into enucleated oocytes (somatic cell nuclear transfer, SCNT) [4]. This finding revealed that cell differentiation from (stem) cells to mature cells is not an irreversible process. Actually, early studies in frogs provided the first experimental evidence that dedifferentiation may occur in non-mammalians (for review, see [5]). The SCNT experiments by Wilmut et al. demonstrated that the enucleated oocyte contains factors that are able to drive cellular reprogramming. Although, these oocyte-specific factors remain so far unidentified, it was the merit of Takahashi and Yamanaka to show that two particular genes, Oct4 and Sox2, which are necessary to maintain pluripotency, are also part of the machinery that is able to induce pluripotency in somatic cells [6]. They demonstrated that overexpression of Oct4 and Sox2 together with two other transcription factors, c-Myc and Klf4, is sufficient to reprogram fibroblast cells into ES-like cells, that have been referred to as induced pluripotent stem cells (iPS cells). Nowadays, with these molecular players of pluripotency induction identified, the analysis of the molecular mechanism underlying factor-induced reprogramming has developed into a major research aim. In particular, the functional contribution of each individual factor is being extensively studied. Recently, c-Myc was found to be dispensable, since Klf4, Sox2 and Oct4 were sufficient to induce iPS formation albeit with decreased efficiency [7, 8]. More recently, Schöler et al. were able to reduce the number of virally transduced reprogramming factors to only one, namely Oct4, by employing neural stem cells that endogenously express Sox2, cMyc and Klf4 [9]. They reasoned that Oct4 is indeed the key factor of pluripotency due to its expression profile during embryonic development. In fact, out of the list of four ‘Yamanaka factors’, Oct4 is the only one that is exclusively expressed in ES cells as well as in primordial germ cells (PGCs). Intriguingly, it has been reported that PGCs can give rise to pluripotent cells (termed embryonic germ cells) when cultured in a simple cocktail of three growth factors [10, 11], which thus could consequently be designated as a ‘No-factor-iPS-protocol’. Taken together, the stem cell factors Oct4 and Sox2 turn out to be essential to drive cellular reprogramming [9, 12, 13]. However, as for SCNT, the molecular mechanism underlying factor-induced reprogramming is poorly understood. Identifying how pluripotency maintenance takes place in ES cells can help us to understand how artificial induction of pluripotency is regulated.
Expression levels of key regulatory factors in ES cells impact on their developmental potential. ES cells are derived from the inner cell mass (ICM) of the blastocyst and exhibit a pluripotent differentiation potential, i.e., they are able to differentiate into any cell type of the body. The transcriptional network of pluripotency is mainly regulated by three factors, Oct4, Sox2 and Nanog. If their expression levels are altered in vitro (overexpression indicated by blue arrows, loss-of function indicated by red arrows) ES cells differentiate toward indicated lineages
Here, we review recent advances in the field of stem cell regulation. A core set of stem cell-specific transcription factors bind cooperatively to promoter regions of several hundred genes involved in the regulation of pluripotency and differentiation. We will summarize the activity of the main factors and discuss their roles in the stem cell machinery. While transcriptional regulation will be one main emphasis of this review, the other focus will be the recently disclosed interconnection between stemness properties and non-coding RNAs. During the last few years, these have become a key focus of scientific attention as a new paradigm of gene expression regulation. Small non-coding RNA molecules are able to influence expression at post-transcriptional level thereby adding another level of complexity to transcriptional networks underlying cellular diversity. We will discuss recent studies deciphering the role of non-coding RNAs in embryonic stem cell biology and their functional interplay with the transcriptional stemness machinery.
Transcriptional control over the stem cell machinery
During recent years, genetic loss- and gain-of-function experiments as well as genome-wide binding analysis revealed the importance of a limited set of transcription factors for the maintenance of stem cell identity. Three transcription factors, Oct4, Sox2 and Nanog (Table 2), turned out to be particularly important, presumably building the top of a transcriptional hierarchy of stemness. Genome-wide DNA-binding analyses have shown that these factors bind cooperatively to promoter regions of several hundred genes involved in regulation of pluripotency and differentiation [14–16]. Thus, understanding the function of these putative master regulators will be instrumental to decipher the stem cell machinery.
Structure and expression pattern of transcription factor Oct4
Oct4 (also known as Oct3 and encoded by the Pou5f1 gene) is a member of the POU-Transcription factor family (Pit, Oct, Unc) [17–19] that contains a bipartite DNA-binding domain designated as POU-domain. It is divided into a POU-specific (POUs) and a POU-homeo domain (POUH), which are connected by a flexible loop. Oct-proteins form a subfamily that bind to an octamer consensus sequence [20]. Members of the Oct-protein-family (Oct1, Oct2, Oct4 and Oct6) show high sequence homologies, especially within regions involved in DNA-binding and POU–POU interaction and are able to form different homo- and heterodimers [21]. They vary in sequences of N- and C-terminal domains that might be responsible for the interaction with different groups of transcription factors and coactivators [22]. In fact, the particular combination of two DNA-binding domains not only confers the specificity of the DNA binding but also provides the flexibility to simultaneously interact with other proteins including regulatory factors.
During early embryonic development, Oct4 is present in toti- and pluripotent cells [18, 19, 23–25]. At the blastocyst stage, Oct4 is expressed in cells of the inner cell mass, but repressed in trophectodermal cells by Cdx2 (Fig. 1) [26]. ES cells, which are derived from the ICM of a blastocyst, seem to maintain this transcriptional fingerprint in vitro. Upon differentiation Oct4 expression in ES cells is downregulated induced by LIF withdrawal or treatment with chemical inducers of differentiation such as retinoic acid. After implantation of the blastocyst, Oct4 expression is restricted to the epiblast and downregulated during gastrulation. Later on, during embryonic development, Oct4 expression is restricted to primordial germ cells [27]. Moreover, Oct4 is expressed in embryonic carcinoma and germ cell lines [17, 24]. The clearly defined expression pattern of Oct4, which is in vivo and in vitro restricted to toti- and pluripotent cells, is indicative of the important role of Oct4 in the maintenance of this developmental particular status. Recently, it has been shown that the Oct4-regulated gene set at the 1- and 2-cell stages of the early embryo is to some extent different from the established network in ES cells [28]. Antisense morpholino oligonucleotide-mediated knock down of Oct4 in 1- to 2-cell embryos showed that in the maternal-embryonic transition Oct4 plays a dual function in post-transcriptional and transcriptional regulation. By this, Oct4 is proposed to affect either directly or indirectly many essential processes during early development, such as chromatin regulation, apoptosis, cell cycle regulation, and signaling [28].
Oct4 as a central player of pluripotency
Several genetic studies corroborate the role of Oct4 as a master regulator of stemness. Oct4 knock-out embryos develop until the blastocyst stage but they do not form a normal ICM [23]. Consequently, ES cells cannot be derived from Oct4-deficient blastocysts. In primordial germ cells, the conditional deletion of Oct4 causes apoptosis [29]. Furthermore, RNAi-mediated knock down of Oct4 in ES cells results in trophectodermal differentiation of the cells (Fig. 1) [30, 31]. Our studies show that a cell-permeable version of Oct4 could rescue the down-regulation of Pou5f1 expression to a large extent [32]. Niwa et al. showed by conditional overexpression of Oct4 that a precise expression level of Oct4 is necessary to sustain pluripotency in ES cells, since about 50% reduced expression already leads to trophectodermal differentiation, whereas a two-fold elevated Oct4 level causes differentiation to primitive endoderm and mesoderm [33] (Fig. 1).
Some studies aimed at assigning Oct4 a potential function in non-embryonic and non-primordial germ cells. It has been reported that Oct4 expression is essential for induction of neural differentiation in ES cells by stromal cell-derived inducing activity (SDIA). Moreover, sustained upregulated Oct4 expression enhanced SDIA-mediated neurogenesis [34]. However, by employing a genetic approach to determine whether Oct4 is important for maintaining multipotency in adult stem cells of several somatic tissues including the intestinal epithelium, bone marrow (hematopoietic and mesenchymal lineages), hair follicle, brain, and liver, it was shown that Pou5f1 ablation revealed no abnormalities in homeostasis or regenerative capacity in these tissues [35]. Some studies assign Oct4 a role in initiating tumorous growth, thereby determining the dark side of stem cells. A report employing a mouse model enabling inducible Oct4-expression in all tissues except brain and testes showed that hyperplasia and invasive tumor formation was induced cell-autonomously in epithelial tissues, suggesting that precursor cells, present in these tissues, are expanded and their differentiation is inhibited [36]. In fact, it has been shown that an aberrant activity of Oct4 may contribute to aneoplastic processes in germ cells [37, 38].
How can a single factor accomplish such a variety of functions? The way in which Oct4 acts on its target genes shows remarkable diversity (Fig. 2a). The N- and C-termini of Oct4 act as transactivators and appear to function redundantly to maintain self renewal [39]. In particular, it could be shown that Oct4 molecules lacking either the N- or C-terminal domain can efficiently replace full-length protein. Even a heterologus transactivation domain of Oct2 fused with the POU domain can sustain self-renewal. This implies that the Oct4 POU domain combined with a proline-rich transactivation domain is sufficient to confer the unique function of Oct4 in ES cells. While Oct4 has predominantly an activating effect on pluripotency related genes like Sox2, Nanog and Pou5f1 itself [40–43], it is indirectly involved in the inhibition of differentiation processes. This is accomplished by activating so-called “repressors of differentiation” like Polycomb group proteins (PcG), zink finger transcription factors, chromatin remodeling factors, and suppressors of signaling [16] (Fig. 3). Post-transcriptional modifications of Oct4 may also result in differential activation potential as has been recently reported. It was found that Oct4 phosphorylation regulate formation of specific Oct4 homodimers with different subsets of target genes [44]. The functional significance of this finding in vivo as well as in ES cells remains to be investigated.
Transcriptional activities and protein–protein interactions of stemness factors (modified from [158]). a Oct4 transcription factor can directly activate gene transcription by binding to octamer sites (first line). Activation may also occur via bridge proteins (second line). Moreover, Oct4 can repress transcription by either direct binding to promoters (third line) or neutralization of activators such as FoxD3 (fourth line). b POU and HMG transcription factors such as Oct4 and Sox2 confer their transcriptional activity often in concert on target genes. Two examples are drawn schematically demonstrating that Oct4 and Sox2 are able to form dissimilar structures of heterodimers that act differently on particular downstream genes. c Nanog is able to block differentiation signals induced by bone morphogenic protein and intracellularly mediated by Smad1 through heterodimerization with Smad1
Oct4/Sox2/Nanog form a core transcriptional network regulating the stem cell machinery. Several hundred target genes are co-occupied by Oct4, Sox2 and Nanog. They can be classified into two groups of downstream genes exerting opposing functions. One group includes genes associated with proliferation, chromatin modification and signaling. On the other hand, the core network activates pathways that lead to the inhibition of differentiation. Polycomb group (PcG) proteins act as transcriptional repressors (for details see text)
The role of Sox2 and its interplay with Oct4
Sox2 is a member of the High Mobility Group (HMG) DNA-binding proteins. HMG proteins are unique in their DNA-binding, since they induce a strong DNA bent through binding to the minor groove of the DNA-helix. By this, HMG proteins endow the chromosome with nuclease sensitivity alluding to an essential role in maintenance and modulation of chromatin structure. Moreover, HMG proteins recruit transcription factors to bind to enhancers [45, 46]. The HMG-domains of Sox (sry-related HMG-box) proteins show highly similar DNA-binding properties, recognizing only a 6- to 7-base pair DNA-sequence. Additionally, Sox proteins contain an activation-/repression-domain that is located at the C-terminus and an interaction domain through which they interact with other partners. Sox proteins are grouped depending on the homologies between their HMG domains; this reaches over 90% within an individual group and about 60% between different groups [46, 47]. Sequence similarities outside the HMG domain determine the sub-grouping of Sox proteins. Sox2, together with Sox1 and Sox3, belongs to the SoxB1 sub-group family [48]. Since the stability of the Sox-DNA-complex is rather low compared to other potent transcription activators (dissociation constant Kd= 10−9−10−8 M), the interaction with coactivators might be instrumental in achieving a stable DNA-binding that results in successful activation of the enhancer, which would explain the cell type-specific action of Sox proteins [49]. For instance, within early embryonic development Sox2 activates target genes by interacting with Oct4. During later developmental stages, with Oct4 not being expressed anymore, Sox2 interacts with Pax6 a major regulator of eye development. The Sox2-Pax6 complex plays an important role in lens development [50].
Sox2 expression is detected in the ICM of mouse blastocysts and further sustains in epiblast [51]. Unlike Oct4, Sox2 also plays a role during later stages of embryonic development and in adult stem cells. After the start of neurogenesis Sox2 is exclusively expressed in the neural plate throughout embryogenesis [51–54], while in the adult organism Sox2 is expressed in neural stem cells of the ventricular zone in the emerging neural tube and their progeny [54–56]. The important role of Sox2 during embryonic development is indicated by the lethality of Sox2-deletion mutants. Murine embryos with homozygously deleted Sox2 do not develop an epiblast, and ES cells lose the ability of proliferation and self-renewal and differentiate into trophectoderm (Fig. 1) [51]. This observation has been recently confirmed by RNAi-mediated knock down and conditional knock out of Sox2 in ES cells [57–60]. The two-fold overexpression of Sox2 has been reported to result in the inhibition of expression of different Sox2 target genes, causing the differentiation of ES cells to neuroectoderm, mesoderm and trophectoderm, but no endoderm [61] (Fig. 1). Thus, similarly to Oct4, a well-balanced precise expression level of Sox2 seems to be important for the maintenance of pluripotency.
POU-proteins are common interaction partners of Sox-factors; together they regulate fundamental processes during development [62]. For instance, a complex between Drosophila Sox2 homolog protein fish hook/Dichaete and the POU-protein Drifter/ventral vienless (Dfr) plays a role in the development of the midline glia [63, 64]. The heterodimerization between Oct4 and Sox2 provides in fact the best characterized example of a POU and SOX partnership (Fig. 2b). Several functional studies have shown that Oct4 and Sox2 directly interact when binding to promoter regions to activate the expression of pluripotency related genes including their own [40, 41, 65], as well as the expression of Nanog, Fgf4, Utf1, and Fbx15 [42, 43, 66–68]. Together with Klf4, they activate the expression of Lefty1 [69]. Interestingly, depending on the target gene, Oct4 and Sox2 exhibit different levels of cooperation [70]. These levels appear to result in gradually different transcriptional activity for downstream genes like Fgf4 and Utf1, depending on the amount of Oct4 and Sox2 proteins present in the cell. Due to a higher level of cooperativity, Oct4 may require less Sox2 to heterodimerize and augment Utf1 activity than is the case for Fgf4 (Fig. 2b). While the function of Oct4 in ES cells has been extensively characterized, the role of Sox2 remained rather undetermined until recently. It has been speculated that Sox2 acts synergistically with Oct4 to activate Oct-Sox enhancers thereby regulating the expression of pluripotency genes including Nanog, Utf1 as well as Oct4 and Sox2 itself (Fig. 2). However, there has been no direct evidence that Sox2 is indeed required for Oct-Sox enhancer activity in ES cells. Masui et al. assessed the role of Sox2 employing an inducible Sox2-null ES cell model. As expected, Sox2-deficient ES cells rapidly differentiate mainly into trophectodermal lineage thereby confirming Sox2 to be indispensable for maintaining ES-cell pluripotency. However, unexpectedly Oct-Sox enhancers remain active even in the absence of Sox2 protein. Moreover, forced expression of Oct4 is able to rescue the pluripotency phenotype of Sox2-null ES cells. These seemingly contradictory observations may be explained by the fact that other Sox proteins present in ES cells, like Sox4, Sox11 and Sox15, could replace the function of Sox2 in coactivation to Oct4-Sox2-regulated gene expression [60].
Expression of Nanog and its cellular activities
The identification of a third key player of pluripotency was based on the functional analysis of the interplay between extrinsic and intrinsic stemness signals of ES cells. The cytokine leukemia inhibitory factor (LIF) is usually indispensable for the cultivation of mouse ES cells [71], and several genetic studies have shown that this extrinsic signal of self-renewal is mediated intracellularly by Stat3 [72]. Neither Oct4 nor Sox2 gain-of-function is able to alter ES cells’ dependence on LIF whereas Nanog overexpression is able to overcome LIF-dependence of mouse ES cells [73, 74]. Nanog expression can be detected in the ICM of murine blastocysts and the epiblast as well as in vitro in ES cells. Upon differentiation, Nanog is generally downregulated; however, it is still expressed in germ cells as well as in tumorous cell lines [73, 75]. Interestingly, in contrast to Oct4 and Sox2, Nanog is not homogeneously distributed between ES cells. Its expression reflects a mosaic-like pattern where Nanog-high populations as well as Nanog-low but Gata6-positive populations can be found within the culture dish. This properly reflects the situation observed in the ICM of the blastocyst at E3.5, where the expression of Nanog and Gata6 resembles a salt-and-pepper pattern [76].
Like Oct4, the Nanog protein contains a homeodomain, exhibiting a structure which it shares with members of the Nk-2 gene family concerning the position of the homeodomain although more than half of the amino acids differ [74]. While there is homology between its orthologs within different species, the Nanog homeoprotein seems to be unique [77]. The most prominent representatives of homeodomain transcription factors can be found within the group of Hox genes, which were discovered in Drosophila melanogaster about 30 years ago [78]. Through their homeodomain, these transcription factors are able to bind and interact with DNA. Hox proteins fulfil the complex tasks like pattern formation [79] and segmentation during morphogenesis [80] through their ability to target a broad range of diverse binding sites. This is accomplished by building heterodimeric complexes with co-factors like Pbx1 and Meis1 and thereby occupying enhancer regions of different sets of target genes (reviewed in [81]). This strategy of binding variety through multimeric diversity is similar to that observed in the cases of Oct4 and Sox2 (Fig. 2).
Transcriptional activities of Nanog and its interaction with other proteins
Mouse Nanog is a three-domain protein consisting of 305 amino acids (aa) encompassing the N-terminal domain (aa 1–aa 95), the homeodomain which is located between aa 96 and aa 155, and the C-terminal part of the protein (aa 156–aa 305) [73, 74]. Thus far, there is only few data on the transcriptional activity of Nanog. Based on luciferase reporter assays in HEK293, NIH3T3, and P19 as well as mouse ES cells, both the N-terminal and the C-terminal domain exhibit transcriptional activity, the C-terminal domain being about 7 times more active [82]. Moreover, this C-terminal domain itself comprises two parts exhibiting strong activation capability. These transactivation domains were designated as CD1 (aa 150–aa 197) and CD2 (aa 248–aa 305). A repetitive sequence in which every fifth residue is a tryptophan, the so-called tryptophan repeat (WR), was identified between aa 198 and aa 247 thus located inbetween CD1 and CD2 [83]. Recent work shed light on potential co-activators or Nanog-protein interactors. Wang et al. identified various binding partners of Nanog, such as Oct4, Nac1, Rif1, Dax1, and Zfp281, by co-immunoprecipitation [84]. First evidence of a putative homodimerization of Nanog was provided by work published by Torres and Watt. Nanog shares homology with Nkx2.5, which is known to form homodimers, and indeed the authors observed Nanog–Nanog interaction by GST pull-down experiments. Furthermore, they reported that Nanog is able to bind NFkB thereby inhibiting pro-differentiation activities and, in cooperation with Stat3, maintaining pluripotency [85]. More recently, Mullin et al. identified the exact position of the dimerization site within the Nanog protein by reporting that Nanog self-associates within the WR [86]. A further study confirmed the concept of a Nanog dimerization through its WR [87]. The authors performed size chromatography of ES cell nuclear extracts revealing putative Nanog dimers. Using different truncated Nanog mutants for co-immunoprecipitation, the position of the dimerization domain was allocated to the WR, correlating well with the data published by Mullin et al. Interestingly, a mutant version of Nanog, incapable of self-dimerizing, did not exhibit cytokine-independent self-renewal in mouse ES cells as judged by colony formation assays. In addition, the authors reported that forced expression of a functional artificial Nanog dimer is able to keep ES cells self-renewing independent of LIF whereas overexpression of the dimerizing-deficient Nanog mutant did result in a loss of typical ES morphology and differentiation. In the presence of LIF, the Nanog dimer as well as the monomeric variant increased the percentage of uniformly growing undifferentiated colonies indicating enhanced self-renewal. Furthermore, the authors showed that dimerization is required for Nanog to be able to interact with other pluripotency network proteins such as Zfp281, Sall4, Dax1, and Zfp198 through co-immunoprecipitation experiments [87]. Besides homo-dimerization Nanog has been reported to physically interact with Smad1 thereby blocking BMP-induced mesoderm differentiation of ES cells [88] (Fig. 2c). It also acts as a direct activator of transcription for the Rex-1 promoter in concert with Sox2 [89]. Moreover, Sall4 was purified in a protein complex together with Nanog and within this complex Nanog positively regulates its own expression as well as that of Sall4 [90]. Pereira et al. reported that Tcf3 is necessary to limit steady-state levels of Nanog mRNA, protein and promoter activity within pluripotent ES cells [91]. In addition, Nanog is able to recruit a repression complex called NODE (Nanog and Oct4 associated deacetylase). In this way, Nanog interacts with Oct4 and the chromatin remodeling machinery since the NODE complex possesses histone deacetylase (HDAC) activity [92].
Genetic analysis of Nanog activity
As outlined above, forced expression of Nanog enables mouse ES cells to self renew in the absence of LIF. Differentiation in response to retinoic acid and 3-methoxybenzamide, two known reagents to promote differentiation, is reduced in Nanog-overexpressing cells. Embryoid body formation was reported to be hampered as well. After Cre-mediated excision of a floxed Nanog transgene the full differentiation potential of these cells was verified by blastocyst injection that resulted in the generation of healthy chimeric mice [73]. There is only few data indicating that the function of Nanog might be conserved between human and mouse ES cells since LIF has been shown anyway not to play a role in the self-renewal of human ES cells [93]. However, it has been reported that enhanced expression of NANOG in human embryonic stem cells enables them to feeder-free growth in unconditioned media [94], indicating that Nanog in both mouse and human ES cells integrates the essential extrinsic stemness signals.
The activity of ectopically overexpressed Nanog in non-ES cells has so far been poorly investigated. Several studies employing cell fusion and iPS cells as model systems assign Nanog a role in the induction of pluripotency in mature cells. Increased levels of Nanog are able to promote pluripotency transfer to the somatic cell genome as shown in cell fusion experiments [95]. Nanog also appears to play a role in the reprogramming process of human somatic cells [96–98]. However, whereas Oct4 and Sox2 have been shown to be essential for cell reprogramming, Nanog is dispensable and seems to have only a promoting effect. A possible role of Nanog during the reprogramming process might be disclosed by experiments of ectopic Nanog expression in somatic cells as well as in stem cells. It has been published that expression of Nanog in NIH 3T3 cells results in enhanced proliferation by promoting cells to enter into S phase [99]. An observation reported by Zhang et al. further supports a potential role of the Nanog protein influencing the cell cycle, as the authors show that NANOG is able to regulate S-phase entry in human embryonic stem cells via transcriptional regulation of CDK6 and CDC25A, two major cell cycle regulatory components [100]. This observation correlates well with that of Mitsui et al. in which they state that ablation of Nanog results in decelerated cell growth in mouse ES cells [74]. Piestun et al. also reported an increased growth rate of Nanog-expressing NIH 3T3 cells. Moreover, this study reported a transformed phenotype of the cells resulting from Nanog expression as judged by cell foci formation [101].
Further insight into the role of Nanog was revealed by loss-of-function studies. Ablation of Nanog results in loss of pluripotency accompanied by an induction of extraembryonic endoderm-associated expression of Gata6 [74] as well as an up-regulation of trophectodermal marker genes like CDX2 (Fig. 1) [102]. Besides endoderm and trophectoderm differentiation, Ivanova et al. showed that Nanog down-regulation by RNAi also induces the expression of markers for epiblast-derived lineages, namely mesoderm, ectoderm and neural crest cells [58].
Although Nanog and Oct4 as well as Sox2 share a high percentage of their target genes, Nanog appears to fulfil a particular and distinct function in the stem cell machinery. In contrast to Oct4, Nanog expression exhibits a fluctuative pattern in morulae during development [103]. Consistently, Nanog was also reported to be heterogeneously expressed in ES cells. Nanog-high populations express further markers of pluripotency such as Oct4, whereas Nanog-low cells also stained positively for primitive endoderm [76]. The heterogenous expression pattern of Nanog in ES cells may indicate different subsets of a pluripotency state. Genetic ablation of Nanog in ES cells provided a clue to the distinct role of Nanog in the stem cell machinery. Chambers et al. genetically deleted Nanog in ES cells by Cre-mediated recombination and reported that Nanog-null ES cells, although prone to differentiation, are still able to self renew. In addition, after aggregation of Nanog-deficient cells with wild-type morulae, they showed that Nanog−/− cells can give rise to post-natal chimeras. Moreover, to investigate whether Nanog-deficient cells can contribute to further development, the authors assessed the persistence of Nanog-null cells during development. They showed that Nanog−/− cells are indeed present in the soma of the genital ridge until E11.5. Consequently, Nanog seems to be required for primordial germ cells to prosecute the germ-cell development program beyond E11.5 [104].In conclusion, these data indicate that, in contrast to Oct4 and Sox2, Nanog is expendable for the maintenance of pluripotency but primarily acts in the establishment and construction of the ICM and germ cell formation. While Oct4 and Sox2 are necessary to direct the stemness program, Nanog appears to be dispensable for the housekeeping machinery of pluripotency.
Genome-wide analysis of stemness factors target genes
Using genome-scale location analysis, sets of target genes for OCT4, SOX2 and NANOG could be identified in the human genome. A substantial amount of target genes was revealed to be co-occupied by these three transcription factors (Fig. 3). The regulatory circuitry seems to consist of autoregulatory and feedforward loops in which first of all the three transcription factors regulate the expression of themselves. Additionally, events associated with the maintenance of stemness-like proliferation, activation of transcription factors and chromatin modifiers as well as stem cell signaling get initiated. Beyond that, factors involved in differentiation processes become downregulated thereby inhibiting embryonic stem cells to acquire an ectodermal, mesodermal or endodermal fate [14] (Fig. 3). In mouse ES cells, Oct4 and Nanog also share a high percentage of their target genes, just like in their human counterparts. Nanog and Oct4 seem to control a cascade of pathways governing pluripotency like REST, self-renewal as seen with nMyc, genome surveillance like in the case of Trp53, as well as cell fate determination [15]. Moreover, Boyer et al. identified the polycomb group (PcG) proteins as transcriptional repressors in mouse ES cells. The PcG repressive complexes PRC1 and PRC2 co-occupy over 500 genes most of which are important developmental regulators [105]. Besides these three seminal publications, further investigation of the core regulatory network in pluripotent cells turns out to be of immense interest [106–110].
Non-coding RNAs and pluripotency maintenance
During the last few years, a new paradigm of genetic regulation has become a key focus of scientific attention. Small non-coding RNA molecules are able to influence expression at post-transcriptional level thereby adding another level of complexity to transcriptional networks underlying cellular diversity [111, 112]. In fact, recent studies revealed exciting novel insights into the role of non-coding RNAs in embryonic stem cell biology. In general, there are several classes of RNAs that do not code for a protein product. Most commonly known are those involved in splicing (ribozymes) and translation (tRNAs). More recently, new classes of non-coding RNAs were identified, changing the classical view of RNAs being just mediators of genetic information. Nowadays, RNA molecules have been assigned an important role in the regulation of biological systems representing highly dynamic nucleotide regulators. Such newly identified non-coding RNAs (ncRNAs) include microRNAs (miRNAs), piRNAs and (endogenous) siRNAs all of which are defined by their way of biogenesis. Endo-siRNAs and piRNAs have been shown to be most abundant in germ cells and are involved among others in anti-viral defense and regulation of “selfish” DNA elements, such as repetitive elements and retrotransposons [113–115]. It is likely that they also play additional roles in the maintenance of stem cell characteristics. However, since so far there are few data on endo-siRNAs and piRNAs in ES cells, we will focus on the rapidly evolving field of miRNAs and their role in maintenance of pluripotency.
Synthesis of micro-RNAs and their mode of action
MiRNAs were first described in the control of timing during larval development of C. elegans [116]. Lee et al. reported that lin-4, a gene essential for normal postembryonic development, does not encode for a protein but was complementary to the 3′ untranslated region (UTR) of the lin-14 transcript. Moreover, it has been shown that the protein level of lin-14 negatively correlates with lin-4 expression, while the mRNA amount of lin-14 is unchanged, indicative of a new level of control for transcriptional homeostasis. Further studies revealed that lin-4 belongs to a highly conserved class of ncRNAs, referred to as microRNAs (miRNAs), which act as an additional regulator of gene expression.
In general, the synthesis of miRNAs begins in the nucleus, where primary miRNAs (pri-miRNAs) are transcribed by RNA-Polymerase II and III (for review, see [117, 112]). The pri-miRNAs contain hairpin structures and are processed by the nuclear microprocessor complex that consists of the RNase III enzyme Drosha and the RNA binding protein dgcr8. The resulting pre-miRNAs that still show the characteristic stem-loops are exported to the cytoplasm where they join with the RNAi pathway. Dicer, another RNase III enzyme, recognizes the pre-miRNAs, cuts off their loop and opts for one strand. The resulting approximately 21 nucleotides are loaded into the RNA-induced silencing complex (RISC), consisting of Dicer, argonaute (Ago) class proteins and target mRNAs bearing partially complementary sites in their 3′-UTRs. The RISC complex subsequently binds to its target mRNAs that are silenced either through direct cleavage or translational repression. For most miRNAs, the nucleotides 2–7 of their sequence are decisive for target recognition. This region, also referred to as ‘seed sequence’, binds to the target-mRNA, unlike the whole miRNA sequence, in a perfect complementary manner consequently exhibiting high homology among related miRNAs. The detailed mechanism of miRNA-mediated gene regulation is a matter of controversy. Substantially, there are two models that also appear to complement one another. The first model relies on the structural relationship of Ago-2 and the translation initiation factor eIF4E. Since Ago-2 is recruited by miRNAs, it has been proposed that silencing takes place during translational initiation by inhibiting the recognition of the cap structure of mRNAs by the translational apparatus [118]. The second model suggests inhibition subsequent to initiation, either by premature ribosome drop-off and/or degradation of the nascent proteins [119, 120]. Finally, the mRNA is sequestrated in the cellular P bodies either as an indirect result of translational inhibition or directly resulting from miRNA targeting [121]. miRNA genes are located throughout the genome in independent transcription units, polycistronic clusters, or within the intron regions of protein-coding genes [112]. Due to their imperfect binding pattern, miRNAs usually target multiple mRNAs. This, in combination with their expression pattern, provides a versatile mechanism for the regulation of developmental processes (Fig. 4).
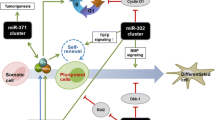
Simplified model of stemness control providing a holistic view that combines the transcriptional network (left) with the activities of non-coding RNAs (right). a Maintenance of stemness. Transcription factors and miRNAs associated with pluripotency balance each other to inhibit differentiation (box at the top in red) and expedite proliferation (box at the bottom in green). b Differentiation. After initiation of differentiation the transcriptional network and miRNA expression change. Tissue-specific transcription factors and miRNAs control accurate differentiation (top; here exemplified by neural differentiation) resulting in a decrease in proliferation and finally exit from the cell cycle (bottom). Activities of ncRNAs exert a weighting and fine-tuning function, e.g., by removing residual counteracting Oct4, Sox2 and Nanog transcripts from the system. Various downstream effector molecules are exemplified. See text for details
Notably, down-regulation might not be the only way how miRNAs regulate gene expression. Place et al. recently described a direct activation of expression by complementary binding of miRNA-373 to its targets’ promoter regions (E-cadherin and cold-shock domain-containing protein C2) [122]. So far, little is known about the RNAa-side of miRNAs but it gives a hint of hidden layers of complexity that remain to be investigated.
Non-coding RNAs and their role in pluripotency maintenance and self renewal
An indication for the involvement of miRNAs in pluripotency control of ES cells has become obvious as miRNA targets were found in the amino acid coding sequence of Nanog, Oct4 and Sox2 [123]. Indeed, miRNAs such as miR-134, miR-296 and miR-470 were found to be up-regulated upon differentiation, leading to transcriptional and morphological changes. Initial functional studies addressing the role of nc-RNAs in stem cells were based on genome-wide deletion of ncRNA-processing enzymes. It was shown that Dicer-Null mouse embryos die around day 7.5 with an almost complete depletion of stem cells, which is consistent with the finding of a premature down-regulation of Oct4 [124]. Furthermore, Dicer-deficient ES cells show a reduced rate of proliferation and severe defects in differentiation [125–127]. Moreover, Dicer-null ES cells also exhibit deficiencies in heterochromatin stability, telomere homeostasis and silencing of transposons [125, 128], which suggests a connection to epigenetic regulation. Taken together, these findings indicate a multifaceted role of ncRNAs during development and homeostasis with complex, context-dependent functionalities. As a consequence, ncRNA activities comprise a contribution to both differentiation and self renewal of ES cells.
miRNAs are involved in cell cycle control
So far, 547 micro RNA genes have been identified in the mouse genome and about half seem to be expressed in ES cells (http://microrna.sanger.ac.uk)[129]. The phenotypes of Dicer- and Drosha-deficient ES cells indicate that miRNAs might be involved in cell cycle control. Murine ES cells show high rates of proliferation, and generation times of about 10 h [130]. The G1 phase is relatively short (~2 h) for the benefit of a high percentage of cells in the S phase. In differentiating cells, the situation is reversed, the fraction of cells in S phase is lower, and the G1 phase elongated. Recently, Wang et al. reported that ES cells deficient for dgcr8, a RNA-binding protein essential for pri-miRNA processing, show a prolonged G1 phase and defects during differentiation [126]. In order to identify particular miRNAs that are involved in cell cycle control, the authors screened a miRNA library for the rescue of the proliferation defect. The authors identified 14 miRNAs, most of which where in the miR-290, miR-302 and miR-17-95 clusters. This finding is in line with other studies reporting these clusters to be the most abundant in ES cells. One of the miRNA candidate targets, cdkn1a (cyclin-dependent kinase inhibitor 1a, also designated as p21), is a gene of particular interest. Cdkn1a inhibits the cyclin-dependent kinase 2 (cdk2) that is constantly expressed during all stages of the ES cell cycle. ES cells are unique in the continuous activity of cyclin:Cdk2 complexes throughout the cell cycle. Cdkn1a inhibits the kinase activity of cyclin:Cdk2 complexes. Thus, the regulation of cdkn1a expression level by miRNAs of the miR-290, miR-302 and miR-17-95 clusters has a direct influence on cdk2 activity. In fact, cdk2 decreases upon differentiation of ES cells and consequently its cell cycle-dependent regulation correlates with differentiation. The implication that miRNAs of the miR-290 cluster positively regulate cdk2 in an indirect manner was supported by miRNA overexpression studies. Coherently, transfection of ES cell with cdkn1a show a similar G1-S delay as observed in Dgcr8 knockouts. Intriguingly, analysis of the seed sequences demonstrated that the identified miRNAs showed a high percentage of redundancy suggesting highly regulated interactions. The introduction of the 14 miRNAs in dgcr8−/− ES cells cells rescued only the ES cell progression delay, the defects in differentiation, however, remained unchanged [126]. Recently published studies confirmed the concept of functional involvement of miRNAs in cell cycle control of stem cells as miR-302b has been reported to directly influence cyclin D2 expression that shows increasing levels upon differentiation [131].
miRNAs and their role in differentiation of stem cells
Certain miRNAs have been shown to regulate differentiation and morphogenesis in a tissue-specific manner [132–134]. For example, during neurogenesis, miRNAs play an important role in the determination of neural lineages from ESCs. Overexpression of miRNA-9 and miRNA-124a in neural precursors lead to a reduction of astrocyte differentiation, whereas knocking-down miR-9 results in a decrease of neurons [135]. Activation of the Stat3 signaling pathway has been shown to inhibit terminal neuronal differentiation [136, 137]. Indeed analysis of Stat3 revealed a negative correlation between the level of its own phosphorylation and miRNA-9 and miRNA-124a expression levels. These findings demonstrate how both miRNAs could indirectly regulate neurogenesis (Fig. 4b). Moreover, target analysis for miRNA-124 identified the small C-terminal domain phosphatase 1 (SCP1) to be posttranscriptionally regulated by miRNA-124. In turn, timely down-regulation of SCP1 seems to be important for induction of neurogenesis in mouse embryos [138].
In addition to tissue-specific miRNAs, there seem to be classes of miRNAs that have a more general role in regulating differentiation and depend rather on the differentiation state than on a specific tissue. Tay et al. [123] recently demonstrated that miRNAs genes are involved in early stages of differentiation. The authors report that upon retinoic acid-induced differentiation, miRNA-134, miRNA-296 and miRNA-470 are up-regulated. These miRNAs directly target the pluripotency factors Oct4, Sox2 and Nanog. This observation indicates a fine-tuning mechanism enabling the complete and rapid disappearance of residual stem cell-specific mRNAs still persisting after transcriptional down-regulation. Notably, the miRNAs 134, 296 and 470 appear to target the mRNA within the coding sequence, contrary to the classic view that binding occurs only in the UTR. However, the specific role of the miRNAs during differentiation remains to be shown. Taking into consideration that miRNAs target a high number of different transcripts, it will be of particular interest whether the identified miRNAs act more generally in weakening the pluripotency network and/or whether they are actively incorporated in certain differentiation pathways.
Another candidate with implications for a more general role in differentiation processes is the let-7 family. Comprising 12 members (let-7a1, a2, a3, b, c, d, e, f1, f2, g, I, and miRNA-98), it is located on 8 different chromosomes [139] and highly conserved among various species [140]. In ES cells, let-7 expression is tightly regulated, among others by lin28. Lin-28 binds to conserved nucleotides within the loop region of the Let-7 precursor and inhibits the processing by Drosha [141]. In mammalian development, let-7 is expressed late and targets a broad set of cell cycle regulators such as cdc25A and cdk6 [142], growth promoting pathways including ras and c-Myc [143, 144] and early embryonic genes such as hmga2, Mlin-41 and IMP-1 [145–148]. This mechanism provides a plausible explanation for the observation that lin-28 is able to promote the induction of pluripotency in somatic cells [98]. When co-expressed together with Oct4, Sox2 and Nanog, lin-28 might mimic the specific activities of Klf4 and c-Myc that represent classical components of the ‘Yamanaka cocktail’ (Oct4, Sox2, Klf4, and c-Myc).
Recently, 12 let-7 regulated oncofetal genes (LOGs) were identified. These are suppressed by let-7 in most adult tissues [147] and expressed in ES cells and various cancers. Among them is the HMG AT-hook 2 (hmga2) gene, encoding a stem cell factor that appears to modulate maintenance of self-renewal of neural stem cells (NSCs) in an age-dependent manner. Investigations employing hmga2 knock-out mice revealed that although this factor is not required for the generation of NSCs, its loss results in defects in proliferation and self-renewal of this cell type. Analyses of cdkn2a (p16INK4a/Arf) expression in NCSs lacking Hmga2 showed a significant overexpression of both tumor suppressor genes [149]. Normally, the cdkn2a locus is activated late in fetal development and a guardian against early neoplastic events such as uncontrolled proliferation.
In conclusion, these observations allude to a model of age-conditioned safeguard in which highly proliferative ES cells and tissue-specific stem cells such as NSCs may self-renew without being addressed as malignant. Aging cells in contrast, where high proliferation may indicate aberrant growth, are tightly controlled. For this, Hmga2 appears to be a key regulator by inhibiting JunB and other targets that presumably activate cdkn2a expression. Intriguingly, Hmga2 is a direct target of Let-7 [147, 149]. Consequently, mi-RNAs such as Let-7 have been hypothesized to be guardians against pluripotency and inappropriate ways toward stemness [150].
How transcriptional networks and non-coding RNAs talk to each other
Cyclin D2 that is a direct target of miR-302 has been shown to negatively correlate with the expression of Oct4 and Nanog. This indicates for an indirect regulation of pluripotency-associated transcription factors by micro-RNAs [131]. The analysis of a human embryonic carcinoma line, whose miR-302 expression pattern strongly resembles that of human ES cells, revealed high correlation of miR-302b and Oct4 expression. Moreover, upon retinoic acid-induced differentiation, the overexpression of miR-302b could rescue Oct4 levels. Further indication of miR-302’s role in the regulation of pluripotency derives from overexpression experiments of the miR-302 cluster in human skin cancer. The resulting cells show an altered cellular phenotype resembling pluripotent human ES cells [151]. It remains to be investigated whether this is due to the balancing of the aberrant transcription network towards pluripotency or whether miR-302s might directly induce pluripotency.
Until recently, there were few data on the molecular and functional interaction between the transcription factor machinery and the non-coding RNA circuitry controlling stemness. Marson et al. reported high-resolution chromatin immunoprecipitation data [152]. The authors identified high-confidence miRNA promoters, based on chromatin modifications and proximity to annotated miRNA-genes, and linked them to binding maps of the transcription factors Oct4, Sox2, Nanog and Tcf3. By this, 55 miRNA genes were identified whose promoters show binding sites for all four factors. Thus, Oct4/Sox2/Nanog together with Tcf3 regulate roughly 20% of miRNA and protein coding genes. Interestingly, a quarter of these miRNAs has been shown to be not actively transcribed in ES cells. Comprehensive epigenetic analysis of the promoters of these miRNA genes showed a tri-methlyation mark at lysine 27 of histone 3 (H3K27me3) and enrichment for polycomb proteins, both of which are characteristic for epigenetically repressed genes. Consequently, it could be demonstrated that these genes were up-regulated in differentiated cells as compared to ES cells.
Other groups have also recently reported indications for an epigenetic connection between miRNAs and pluripotency factors. Benetti et al. and Sinkkonen et al. described methylation defects in Dicer-deficient ES cells, concomitant with increased telomere recombination and elongation [128] and insufficient silencing of the Oct4 promoter during differentiation [153]. Again, the miR-290 cluster turned out to be of particular importance as it was identified as targeting the retinoblastoma-like 2 protein (Rbl2), a transcriptional repressor. Corresponding to high Rbl2 levels in Dicer-null ES cells, the expression of DNA-methyltransferases (Dnmt), particularly the de-novo Dnmts 3a and 3b, declined. The methylation defects could be rescued either by expression of Dnmts or by transfection of miR-290s [153], suggesting that miR-290 indirectly regulates epigenetic modification through a transcriptional repressor. Interestingly, the epigenetic defects only concerned DNA-methylation whereas repressive histone modification remained stable.
Concluding remarks and outlook
As we have described, studies in the last decade have enabled researchers to get a comprehensive understanding of how stemness maintenance in pluripotent cells is regulated. In conclusion, the control of stemness properties in ES cells appears complex (Fig. 4). ES cells show highly accessible chromatin that makes them prone to transcriptional leakiness and consequently holds risk for chaotic or even dangerous resonance effects that might result in apoptosis or malignances. This explains the need for buffering the stemness machinery at different levels through transcription factors, miRNAs, and epigenetic marks. This is particularly important for genes whose inappropriate expression results in undirected differentiation or malignant proliferation. The groundbreaking work by Takahashi and Yamanaka [6] demonstrated that a rather marginal manipulation of the stemness network is sufficient to induce pluripotency in somatic cells. Although the underlying mechanism remains elusive, researchers are beginning to understand how transcriptional and miRNA networks are talking to each other in order to decide a stem cell’s fate. Initially, four virally transfected factors, Oct4, Sox2, Klf4, and c-Myc, were reported to be sufficient to induce pluripotency in fibroblasts [6]. A further study reported that reprogramming of human ES cells is feasible by another combination of factors, namely Oct4, Sox2, Nanog, and LIN28 [98]. Taking these findings together, Oct4 and Sox2 emerge as the two key regulators and inducers of pluripotency. Nanog appears to play a role only in promoting the induction of pluripotency, whereas the miRNA-specific activities of LIN28 might mimic the overexpression of Klf4 and c-Myc.
The derivation of iPS cells is a rather inefficient process typically enabling only 1 in 1,000 recipient cells to be reprogrammed. Several explanations may account for this low efficiency. Due to the viral mode of factor delivery, other factors might be involved in reprogramming that are activated through random insertion of the transgenic viral genome. Recent work, however, demonstrated that stable viral integration is not required to generate iPS cells when non-integrating adenoviruses or plasmids are used [154–156]. Additionally, deletable viral constructs and non-viral transponsons have been shown to induce pluripotency in mouse and human ES cells [156, 157]. Thus, insertional mutagenesis fails to explain the overall low efficiency of transcription factor-induced reprogramming. One possible explanation is that iPS generation requires rare stochastic events. In contrast to well-balanced ES cells, some somatic cell types might be more accessible to respond to transcriptional noise at least in the artificial context of cell culture. Rare cells might escape the transcriptional balance determining the cellular phenotype, and then a comparably weak trigger like Oct4-overexpression might be able to induce a dramatic transcriptional switch. The imbalance might be caused by factors like c-Myc, chemicals interfering with key signaling pathways, or any cell culture condition that increases transcriptional noise.
Stem cells exhibit very special cellular properties by combining the ability to renew themselves and to differentiate into specialized cell types. Thus, stem cells can be used for cell replacement therapies and for studying differentiation processes in vitro. Although ES cells as well as iPS cells represent a virtually unlimited source of highly versatile cells, clinical application is still in its infancy. Besides targeted differentiation into the wanted cellular phenotype, the major limitation lies in gaining sufficiently tight control over self-renewal activity after transplantation. Adult or somatic stem cells by contrast that can be derived from multiple organs of the adult organism, such as bone marrow, skin and brain, have already been successfully applied in the clinics. In fact, from a therapeutic point of view, pluripotency is not a desirable criterion for cell replacement strategies. A reliable strategy to induce a multipotent or even unipotent state might even be sufficient and much safer for clinical applications. Future studies will show whether or not researchers will be able to gain sufficient insight into the stem cell machinery and learn to precisely manipulate it in order to determine any given cell type in the plastic dish.
References
Chambers I, Smith A (2004) Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 23:7150–7160
Naveiras O, Daley GQ (2006) Stem cells and their niche: a matter of fate. Cell Mol Life Sci 63:760–766
Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156
Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385:810–813
Gurdon JB, Melton DA (2008) Nuclear reprogramming in cells. Science 322:1811–1815
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Wernig M, Meissner A, Cassady JP, Jaenisch R (2008) c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2:10–12
Okita K, Ichisaka T, Yamanaka S (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317
Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR (2009) Oct4-induced pluripotency in adult neural stem cells. Cell 136:411–419
Matsui Y, Zsebo K, Hogan BL (1992) Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70:841–847
Resnick JL, Bixler LS, Cheng L, Donovan PJ (1992) Long-term proliferation of mouse primordial germ cells in culture. Nature 359:550–551
Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR (2008) Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454:646–650
Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA (2008) Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26:1269–1275
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38:431–440
Sharov AA, Masui S, Sharova LV, Piao Y, Aiba K, Matoba R, Xin L, Niwa H, Ko MS (2008) Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics 9:269
Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H (1990) A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60:461–472
Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM (1990) A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345:686–692
Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P (1990) New type of POU domain in germ line-specific protein Oct-4. Nature 344:435–439
Herr W, Cleary MA (1995) The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev 9:1679–1693
Tomilin A, Remenyi A, Lins K, Bak H, Leidel S, Vriend G, Wilmanns M, Scholer HR (2000) Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 103:853–864
Remenyi A, Scholer HR, Wilmanns M (2004) Combinatorial control of gene expression. Nat Struct Mol Biol 11:812–815
Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379–391
Palmieri SL, Peter W, Hess H, Scholer HR (1994) Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol 166:259–267
Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR (1996) Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122:881–894
Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J (2005) Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917–929
Pesce M, Scholer HR (2000) Oct-4: control of totipotency and germline determination. Mol Reprod Dev 55:452–457
Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, Zuo E, Eckart M, Reijo Pera RA, Wong WH, Yao MW (2008) A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE 3:e4109
Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin KJ, Scholer HR, Tomilin A (2004) Oct4 is required for primordial germ cell survival. EMBO Rep 5:1078–1083
Hay DC, Sutherland L, Clark J, Burdon T (2004) Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22:225–235
Hough SR, Clements I, Welch PJ, Wiederholt KA (2006) Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of OCT4 and Nanog. Stem Cells 24:1467–1475
Bosnali M, Edenhofer F (2008) Generation of transducible versions of transcription factors Oct4 and Sox2. Biol Chem 389:851–861
Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24:372–376
Shimozaki K, Nakashima K, Niwa H, Taga T (2003) Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development 130:2505–2512
Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R (2007) Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 1:403–415
Hochedlinger K, Yamada Y, Beard C, Jaenisch R (2005) Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121:465–477
Gidekel S, Pizov G, Bergman Y, Pikarsky E (2003) Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell 4:361–370
Looijenga LH, Stoop H, de Leeuw HP, de Gouveia Brazao CA, Gillis AJ, van Roozendaal KE, van Zoelen EJ, Weber RF, Wolffenbuttel KP, van Dekken H, Honecker F, Bokemeyer C, Perlman EJ, Schneider DT, Kononen J, Sauter G, Oosterhuis. JW (2003) POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res 63:2244–2250
Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J (2002) Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol 22:1526–1536
Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F (2005) Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem 280:5307–5317
Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25:6031–6046
Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol 25:2475–2485
Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P (2005) Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem 280:24731–24737
Saxe JP, Tomilin A, Scholer HR, Plath K, Huang J (2009) Post-translational regulation of Oct4 transcriptional activity. PLoS ONE 4:e4467
Grosschedl R, Giese K, Pagel J (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 10:94–100
Wegner M (1999) From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res 27:1409–1420
Pevny LH, Lovell-Badge R (1997) Sox genes find their feet. Curr Opin Genet Dev 7:338–344
Uchikawa M, Kamachi Y, Kondoh H (1999) Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev 84:103–120
Kamachi Y, Uchikawa M, Kondoh H (2000) Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet 16:182–187
Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H (2001) Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev 15:1272–1286
Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17:126–140
Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT (1995) Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev 49:23–36
Wood HB, Episkopou V (1999) Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev 86:197–201
Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK (2000) Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development 127:2367–2382
Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK (2004) Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131:3805–3819
Graham V, Khudyakov J, Ellis P, Pevny L (2003) SOX2 functions to maintain neural progenitor identity. Neuron 39:749–765
Fong H, Hohenstein KA, Donovan PJ (2008) Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26:1931–1938
Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR (2006) Dissecting self-renewal in stem cells with RNA interference. Nature 442:533–538
Li J, Pan G, Cui K, Liu Y, Xu S, Pei D (2007) A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J Biol Chem 282:19481–19492
Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9:625–635
Kopp JL, Ormsbee BD, Desler M, Rizzino A (2008) Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26:903–911
Dailey L, Basilico C (2001) Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J Cell Physiol 186:315–328
Ma Y, Certel K, Gao Y, Niemitz E, Mosher J, Mukherjee A, Mutsuddi M, Huseinovic N, Crews ST, Johnson WA, Nambu JR (2000) Functional interactions between Drosophila bHLH/PAS, Sox, and POU transcription factors regulate CNS midline expression of the slit gene. J Neurosci 20:4596–4605
Soriano NS, Russell S (1998) The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development 125:3989–3996
Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A (2002) Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res 30:3202–3213
Yuan H, Corbi N, Basilico C, Dailey L (1995) Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev 9:2635–2645
Nishimoto M, Fukushima A, Okuda A, Muramatsu M (1999) The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol 19:5453–5465
Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M, Niwa H, Yamanaka S (2003) Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol 23:2699–2708
Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS, Niwa H (2006) Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol 26:7772–7782
Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M (2003) Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev 17:2048–2059
Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D (1988) Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336:688–690
Boeuf H, Hauss C, Graeve FD, Baran N, Kedinger C (1997) Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J Cell Biol 138:1207–1217
Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643–655
Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631–642
Hart AH, Hartley L, Ibrahim M, Robb L (2004) Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn 230:187–198
Singh AM, Hamazaki T, Hankowski KE, Terada N (2007) A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 25:2534–2542
Pan G, Thomson JA (2007) Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res 17:42–49
Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276:565–570
Murphy P, Davidson DR, Hill RE (1989) Segment-specific expression of a homoeobox-containing gene in the mouse hindbrain. Nature 341:156–159
Lohmann I, McGinnis N, Bodmer M, McGinnis W (2002) The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell 110:457–466
Svingen T, Tonissen KF (2006) Hox transcription factors and their elusive mammalian gene targets. Heredity 97:88–96
Pan GJ, Pei DQ (2003) Identification of two distinct transactivation domains in the pluripotency sustaining factor nanog. Cell Res 13:499–502
Pan G, Pei D (2005) The stem cell pluripotency factor NANOG activates transcription with two unusually potent subdomains at its C terminus. J Biol Chem 280:1401–1407
Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444:364–368
Torres J, Watt FM (2008) Nanog maintains pluripotency of mouse embryonic stem cells by inhibiting NFkappaB and cooperating with Stat3. Nat Cell Biol 10:194–201
Mullin NP, Yates A, Rowe AJ, Nijmeijer B, Colby D, Barlow PN, Walkinshaw MD, Chambers I (2008) The pluripotency rheostat Nanog functions as a dimer. Biochem J 411:227–231
Wang J, Levasseur DN, Orkin SH (2008) Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA 105:6326–6331
Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodriguez-Esteban C, Izpisua Belmonte JC (2006) Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci USA 103:10294–10299
Shi W, Wang H, Pan G, Geng Y, Guo Y, Pei D (2006) Regulation of the pluripotency marker Rex-1 by Nanog and Sox2. J Biol Chem 281:23319–23325
Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, Ng HH (2006) Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem 281:24090–24094
Pereira L, Yi F, Merrill BJ (2006) Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol 26:7479–7491
Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10:731–739
Daheron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, Itskovitz-Eldor J, Daley GQ (2004) LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells 22:770–778
Darr H, Mayshar Y, Benvenisty N (2006) Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development 133:1193–1201
Silva J, Chambers I, Pollard S, Smith A (2006) Nanog promotes transfer of pluripotency after cell fusion. Nature 441:997–1001
Ebert AD, Yu J, Rose FF Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN (2009) Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457:277–280
Liao J, Wu Z, Wang Y, Cheng L, Cui C, Gao Y, Chen T, Rao L, Chen S, Jia N, Dai H, Xin S, Kang J, Pei G, Xiao L (2008) Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res 18:600–603
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson. JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920
Zhang J, Wang X, Chen B, Suo G, Zhao Y, Duan Z, Dai J (2005) Expression of Nanog gene promotes NIH3T3 cell proliferation. Biochem Biophys Res Commun 338:1098–1102
Zhang X, Neganova I, Przyborski S, Yang C, Cooke M, Atkinson SP, Anyfantis G, Fenyk S, Keith WN, Hoare SF, Hughes O, Strachan T, Stojkovic M, Hinds PW, Armstrong L, Lako M (2009) A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol 184:67–82
Piestun D, Kochupurakkal BS, Jacob-Hirsch J, Zeligson S, Koudritsky M, Domany E, Amariglio N, Rechavi G, Givol D (2006) Nanog transforms NIH3T3 cells and targets cell-type restricted genes. Biochem Biophys Res Commun 343:279–285
Hyslop L, Stojkovic M, Armstrong L, Walter T, Stojkovic P, Przyborski S, Herbert M, Murdoch A, Strachan T, Lako M (2005) Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23:1035–1043
Dietrich JE, Hiiragi T (2007) Stochastic patterning in the mouse pre-implantation embryo. Development 134:4219–4231
Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450:1230–1234
Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353
Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA (2008) Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22:746–755
Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH (2008) A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10:353–360
Kim J, Chu J, Shen X, Wang J, Orkin SH (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132:1049–1061
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301–313
Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H, Lim B (2008) T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells 26:2019–2031
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev 22:2773–2785
Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H (2008) Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453:793–797
Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC (2008) The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453:803–806
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10:126–139
Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z (2007) An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129:1141–1151
Petersen CP, Bordeleau ME, Pelletier J, Sharp PA (2006) Short RNAs repress translation after initiation in mammalian cells. Mol Cell 21:533–542
Nottrott S, Simard MJ, Richter JD (2006) Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 13:1108–1114
Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20:515–524
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA 105:1608–1613
Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455:1124–1128
Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ (2003) Dicer is essential for mouse development. Nat Genet 35:215–217
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40:1478–1483
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 102:12135–12140
Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA (2008) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15:268–279
Wang Y, Keys DN, Au-Young JK, Chen C (2009) MicroRNAs in embryonic stem cells. J Cell Physiol 218:251–255
Burdon T, Smith A, Savatier P (2002) Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 12:432–438
Lee NS, Kim JS, Cho WJ, Lee MR, Steiner R, Gompers A, Ling D, Zhang J, Strom P, Behlke M, Moon SH, Salvaterra PM, Jove R, Kim KS (2008) miR-302b maintains “stemness” of human embryonal carcinoma cells by post-transcriptional regulation of Cyclin D2 expression. Biochem Biophys Res Commun 377:434–440
Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86
Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT (2004) MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 36:1079–1083
Yekta S, Shih IH, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594–596
Krichevsky AM, Sonntag KC, Isacson O, Kosik KS (2006) Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24:857–864
Moon C, Yoo JY, Matarazzo V, Sung YK, Kim EJ, Ronnett GV (2002) Leukemia inhibitory factor inhibits neuronal terminal differentiation through STAT3 activation. Proc Natl Acad Sci USA 99:9015–9020
Gu F, Hata R, Ma YJ, Tanaka J, Mitsuda N, Kumon Y, Hanakawa Y, Hashimoto K, Nakajima K, Sakanaka M (2005) Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J Neurosci Res 81:163–171
Visvanathan J, Lee S, Lee B, Lee JW, Lee SK (2007) The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21:744–749
Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME (2007) Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle 6:2585–2590
Caygill EE, Johnston LA (2008) Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 18:943–950
Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14:1539–1549
Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ (2007) The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67:7713–7722
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647
Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ (2007) MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67:9762–9770
Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315:1576–1579
Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME (2007) Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 104:11400–11405
Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME (2008) Identification of let-7-regulated oncofetal genes. Cancer Res 68:2587–2591
Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ (2008) The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7:3935–3942
Nishino J, Kim I, Chada K, Morrison SJ (2008) Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135:227–239
Peter ME (2009) Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 8:843–852
Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY (2008) Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. Rna 14:2115–2124
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134:521–533
Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15:259–267
Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K (2008) Induced pluripotent stem cells generated without viral integration. Science 322:945–949
Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S (2008) Generation of mouse induced pluripotent stem cells without viral vectors. Science 322:949–953
Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K (2009) Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458:771–775
Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A (2009) piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458:766–770
Pan GJ, Chang ZY, Scholer HR, Pei D (2002) Stem cell pluripotency and transcription factor Oct4. Cell Res 12:321–329
Acknowledgments
We thank Oliver Brüstle and all members of the Stem Cell Engineering Group, University of Bonn, for support and valuable discussions and the reviewers for their critical reading and helpful suggestions. We do apologize to those whose work was not cited or insufficiently cited because of space limitations. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), Research Priority Program SPP1356, Pluripotency and Cellular Reprogramming (ED79/1-1); the German Ministry of Education and Research (BMBF, 01 GN 0813); and the BONFOR Research Program of the Medical Faculty of the University of Bonn.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Bosnali, B. Münst and M. Thier have contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bosnali, M., Münst, B., Thier, M. et al. Deciphering the stem cell machinery as a basis for understanding the molecular mechanism underlying reprogramming. Cell. Mol. Life Sci. 66, 3403–3420 (2009). https://doi.org/10.1007/s00018-009-0095-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0095-2