Abstract
Background
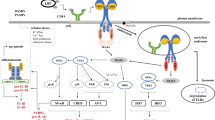
The Notch signaling pathway is a cell signaling system that is conserved in a variety of eukaryotes. Overall, Notch receptors and their ligands are single-pass transmembrane proteins, which often require cell–cell interactions and proteolytic processing to promote signaling. Since its discovery, it has been the subject of extensive research that revealed its importance in several cellular mechanisms, including cell fate determination, hematopoiesis, tissue self-renewal, proliferation, and apoptosis during embryogenesis. Many studies have described the influence of the Notch pathway in modulating the innate and adaptive immune systems.
Methods
We analyzed the literature on the role of the Notch pathway in regulating immune responses during infections, aiming to discuss the importance of establishing a Notch signaling pathway-based approach for predicting the outcome of infectious diseases.
Conclusion
In this review, we present an overview of evidence that demonstrates the direct and indirect effects of interaction between the Notch signaling pathway and the immune responses against bacterial, viral, fungal, and parasitic infections, as well as the importance of this pathway to predict the outcome of infectious diseases.



Similar content being viewed by others
References
Morgan TH, Bridges CB. Sex-linked inheritance in Drosophila. Carnegie institution of Washington; 1916.
Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol (Elsevier). 2000;228:151–65.
Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood Am Soc Hematol. 2006;107:2223–33.
Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky A V, Degnan BM, et al. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. BioMed Central. 2009;9:249.
Fitzgerald K, Wilkinson HA, Greenwald I. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development (Oxford University Press for The Company of Biologists Limited). 1993;119:1019–27.
Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33.
Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol (Nature Publishing Group). 2006;7:678.
de Celis JF, Bray SJ. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development (The Company of Biologists Ltd). 2000;127:1291–302.
Cordle J, Redfield C, Stacey M, Van Der Merwe PA, Willis AC, Champion BR, et al. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J Biol Chem. 2008;283:11785–93.
Raya Á, Kawakami Y, Rodríguez-Esteban C, Ibañes M, Rasskin-Gutman D, Rodríguez-León J, et al. Notch activity acts as a sensor for extracellular calcium during vertebrate left – right determination. Nature. 2004;427:121–8.
Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol Am Soc Microbiol. 2006;26:4642–51.
D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–67.
Chillakuri CR, Sheppard D, Lea SM, Handford PA. Notch receptor–ligand binding and activation: Insights from molecular studies. Semin Cell Dev Biol. (Elsevier) 2012; 421–8.
Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell (Elsevier). 2009;137:216–33.
Pintar A, De Biasio A, Popovic M, Ivanova N, Pongor S. The intracellular region of Notch ligands: does the tail make the difference? Biol Direct. 2007;2:1–13.
Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008;6:e196.
Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y, Takai Y. MAGI1 recruits Dll1 to cadherin-based adherens junctions and stabilizes it on the cell surface. J Biol Chem United States. 2005;280:26499–507.
Gazave E, Guillou A, Balavoine G. History of a prolific family: the Hes/Hey-related genes of the annelid Platynereis. Evodevo. BioMed Central 2014;5:29.
Katoh M, Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol (Spandidos Publications). 2007;31:461–6.
Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SHE, et al. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur J Immunol (Wiley Online Library). 2008;38:174–83.
Foldi J, Chung AY, Xu H, Zhu J, Outtz HH, Kitajewski J, et al. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J Immunol Am Assoc Immnol. 2010;185:5023–31.
Fung E, Tang S-MT, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, et al. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation (Am Heart Assoc) 2007;115:2948–56.
Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell (Springer). 2016;7:159–74.
Ito T, Connett JM, Kunkel SL, Matsukawa A. Notch system in the linkage of innate and adaptive immunity. J Leukoc Biol (internet). 2012;92:59–65. https://doi.org/10.1189/jlb.1011529.
Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity (Elsevier). 2010;32:14–27.
Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99.
Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–26.
Maekawa Y, Ishifune C, Tsukumo S, Hozumi K, Yagita H, Yasutomo K. Notch controls the survival of memory CD4+ T cells by regulating glucose uptake. Nat Med. (Nature Publishing Group) 2015;21:55.
Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol Annual Reviews. 2005;23:945–74.
Cheng P, Gabrilovich D. Notch signaling in differentiation and function of dendritic cells. Immunol Res Springer. 2008;41:1–14.
Koyanagi A, Sekine C, Yagita H. Expression of Notch receptors and ligands on immature and mature T cells. Biochem Biophys Res Commun (Elsevier). 2012;418:799–805.
Schaller MA, Allen RM, Kimura S, Day CL, Kunkel SL. Systemic expression of Notch ligand delta-like 4 during mycobacterial infection alters the T cell immune response. Front Immunol. 2016;7:1–11.
Yamaguchi E, Chiba S, Kumano K, Kunisato A, Takahashi T, Takahashi T, et al. Expression of Notch ligands, Jagged 1, 2 and Delta1 in antigen presenting cells in mice. Immunol Lett. 2002;81:59–64.
Fasnacht N, Huang H-Y, Koch U, Favre S, Auderset F, Chai Q, et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med. (The Rockefeller University Press) 2014;211:2265–79.
Hildebrand D, Uhle F, Sahin D, Krauser U, Weigand MA, Heeg K. The interplay of Notch signaling and STAT3 in TLR-activated human primary monocytes. Front Cell Infect Microbiol. (Frontiers Media SA) 2018;8.
Nikolic N, Jakovljevic A, Carkic J, Beljic-Ivanovic K, Miletic M, Soldatovic I, et al. Notch signaling pathway in apical periodontitis: correlation with bone resorption regulators and proinflammatory cytokines. J Endod (Elsevier). 2019;45:123–8.
Al-Attar A, Alimova Y, Kirakodu S, Kozal A, Novak MJ, Stromberg AJ, et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA 2-IIA. Mucosal Immunol. (Nature Publishing Group) 2018;11:1047.
World Health Organization. Tuberculosis (internet). Tuberculosis. 2020. https://www.who.int/news-room/fact-sheets/detail/tuberculosis#:~:text=Keyfacts,withtuberculosis(TB)worldwide.
Narayana Y, Balaji KN. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J Biol Chem. 2008;283:12501–11.
Bansal K, Narayana Y, Patil SA, Balaji KN. M. bovis BCG induced expression of COX-2 involves nitric oxide-dependent and -independent signaling pathways. J Leukoc Biol. 2009;85:804–16. https://doi.org/10.1189/jlb.0908561
Kapoor N, Narayana Y, Patil SA, Balaji KN. Nitric oxide is involved in Mycobacterium bovis bacillus Calmette-Guérin-activated Jagged1 and Notch1 signaling. J Immunol. 2010;184:3117–26.
Palaga T, Ratanabunyong S, Pattarakankul T, Sangphech N, Wongchana W, Hadae Y, et al. Notch signaling regulates expression of Mcl-1 and apoptosis in PPD-treated macrophages. Cell Mol Immunol (Nature Publishing Group). 2013;10:444–52. https://doi.org/10.1038/cmi.2013.22.
Ito T, Schaller M, Hogaboam CM, Standiford TJ, Sandor M, Lukacs NW, et al. TLR9 regulates the mycobacteria-elicited pulmonary granulomatous immune response in mice through DC-derived Notch ligand delta-like 4. J Clin Invest. 2009;119:33–46.
Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J Immunol Am Assoc Immnol. 2009;182:7381–8.
Castro RC, Zambuzi FA, Fontanari C, de Morais FR, Bollela VR, Kunkel SL, et al. NOTCH1 and DLL4 are involved in the human tuberculosis progression and immune response activation. Tuberculosis (Elsevier). 2020;1:101980.
Li Q, Zhang H, Yu L, Wu C, Luo X, Sun H, et al. Down-regulation of Notch signaling pathway reverses the Th1/th2 imbalance in tuberculosis patients. Int Immunopharmacol (Elsevier). 2018;54:24–32.
da Silva MV, Tiburcio MGS, Machado JR, Silva DAA, Rodrigues DBR, Rodrigues V, et al. Complexity and controversies over the cytokine profiles of T helper cell subpopulations in tuberculosis. J Immunol Res (Hindawi Publishing Corporation) 2015;2015:639107. https://pubmed.ncbi.nlm.nih.gov/26495323
Domingo-Gonzalez R, Prince O, Cooper A, Khader S. Cytokines and chemokines in Mycobacterium tuberculosis infection. Microbiol Spectr. (NIH Public Access) 2016;4.
Dua B, Upadhyay R, Natrajan M, Arora M, Narayanaswamy BK, Joshi B. Notch signaling induces lymphoproliferation, T helper cell activation and Th1/Th2 differentiation in leprosy. Immunol Lett (Elsevier). 2019;207:6–16.
Liu T, He W, Li Y (2016) Helicobacter pylori infection of gastric epithelial cells affects NOTCH pathway in vitro. Dig Dis Sci. (Springer US). 2016.
Ehrlichia chaffeensis TRP120 activates canonical Notch signaling to downregulate TLR2/4 expression and promote intracellular survival. 2016;7:1–15.
Schaller MA, Neupane R, Rudd BD, Kunkel SL, Kallal LE, Lincoln P, et al. Notch ligand delta-like 4 regulates disease pathogenesis during respiratory viral infections by modulating Th2 cytokines. 2007;204:2925–34.
Ting H-A, de Almeida Nagata D, Rasky AJ, Malinczak C-A, Maillard IP, Schaller MA, et al. Notch ligand delta-like 4 induces epigenetic regulation of Treg cell differentiation and function in viral infection. Mucosal Immunol. (Nature Publishing Group) 2018;11:1524.
Ito T, Allen RM, Carson WF, Schaller M, Cavassani KA, Hogaboam CM, et al. The critical role of Notch ligand Delta-like 1 in the pathogenesis of influenza A virus (H1N1) infection. PLoS Pathog. 2011;7:e1002341. http://www.ncbi.nlm.nih.gov/pubmed/22072963%5Cnpubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3207886.
Backer RA, Helbig C, Gentek R, Kent A, Laidlaw BJ, Dominguez CX, et al. A central role for Notch in effector CD8(+) T cell differentiation. Nat Immunol. 2014;15:1143–51. https://pubmed.ncbi.nlm.nih.gov/25344724
Kongkavitoon P, Tangkijvanich P, Hirankarn N. Hepatitis B virus HBx activates Notch signaling via Delta-like 4/Notch1 in hepatocellular carcinoma. 2016;1–15.
Yang S-L, Ren Q-G, Zhang T, Pan X, Wen L, Hu J-L, et al. Hepatitis B virus X protein and hypoxia-inducible factor-1α stimulate Notch gene expression in liver cancer cells. Oncol Rep (Spandidos Publications). 2017;37:348–56.
Wei X, Wang J, Hao C, Yang X, Wang L, Lian J. Notch signaling contributes to liver inflammation by regulation of interleukin-22-producing cells in hepatitis B virus infection. 2016;6:1–10.
Jiang B-C, Liu X, Liu X-H, Li Z-S-N, Zhu G-Z. Notch Signaling regulates circulating T helper 22 cells in patients with chronic hepatitis C. Viral Immunol. (Mary Ann Liebert, Inc) 2017;30:522–32.
Mukherjee S, Akbar I, Kumari B, Vrati S, Basu A, Banerjee A. Japanese Encephalitis Virus-induced let-7a/b interacted with the NOTCH-TLR7 pathway in microglia and facilitated neuronal death via caspase activation. J Neurochem Engl. 2019;149:518–34.
Zhang L, Li H, Hai Y, Yin W, Li W, Zheng B, et al. CpG in combination with an inhibitor of Notch signaling suppresses formalin-inactivated respiratory syncytial virus-enhanced airway hyperresponsiveness and inflammation by inhibiting Th17 memory responses and promoting tissue-resident memory cells in lung. J Virol Am Soc Microbiol. 2017;91:e02111-e2116.
Mas D, Gilberto R, Kaihami H, Pereira G, Fogac R, Almeida D, et al. The critical role of Notch1-TLR 4 signaling in the inflammatory and fungicidal activity of macrophages against Paracoccidioides brasiliensis strain Pb18. 2017;797–807.
Jannuzzi G, de Almeida J, dos Santos S, de Almeida S, Ferreira K. Notch signaling is required for dendritic cell maturation and T cell expansion in paracoccidioidomycosis. Mycopathologia. 2018;3:739–49.
Neal LM, Qiu Y, Chung J, Xing E, Cho W, Malachowski AN, et al. T cell-restricted Notch signaling contributes to pulmonary Th1 and Th2 immunity during Cryptococcus neoformans. Infection. 2019.
Xiong Y, Lingrel JB, Wüthrich M, Klein BS, Vasudevan NT, Jain MK, et al. Transcription factor KLF2 in dendritic cells downregulates Th2 programming via the HIF-1α/Jagged2/Notch axis. Cassone A, editor. MBio. 2016;7:e00436–16.
Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, et al. Notch signaling is an important regulator of type 2 immunity. 2005;202:1037–42.
Krawczyk CM, Sun J, Pearce EJ, Cells D, Krawczyk CM, Sun J, et al. Th2 differentiation is unaffected by Jagged2 expression on. 2015.
Acknowledgements
RCC and FAZ were supported by CAPES, Finance Code 001. FGF was supported by FAPESP (2018/15066-0).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no reported conflicts of interest.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Castro, R.C., Gonçales, R.A., Zambuzi, F.A. et al. Notch signaling pathway in infectious diseases: role in the regulation of immune response. Inflamm. Res. 70, 261–274 (2021). https://doi.org/10.1007/s00011-021-01442-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-021-01442-5