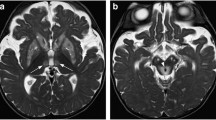
Abstract
The Basal Ganglia (BG) is a small brain structure that plays a significant role in the Parkinson’s disease (PD) pathogenesis. Since it provides detailed quantitative information for the PD progression, positron emission tomography (PET) is an ideal tool for the detection of the amount changes in this region. To make these images easy to analyze and more meaningful, it is crucial to modify their representation and clarify the image through segmentation. It is critical to locate the BG region so that it can be easily examined to detect its size and shape for progression diagnosis. In this paper, an exhaustive study of BG segmentation from PET images is performed through different segmentation techniques, namely binary thresholding, truncate thresholding, threshold to zero, Otsu thresholding, and clustering combined with binary thresholding. These techniques have been tested onto 110 PET images and evaluated with their corresponding ground truth which were previously segmented manually. We evaluated the segmentation performance using two metrics Dice Similarity Coefficient (DSC) and mean Intersection over Union (mIoU). The obtained results indicate that binary thresholding technique outperforms other segmentation techniques and reached higher performance using 150 as threshold with DSC of 0.7701 and mIoU of 0.6394.



Similar content being viewed by others
Data availability
The data used in this paper was obtained from the Parkinson’s Progressive Marker Initiative (PPMI) database (https://www.ppmi-info.org/). These restrictions must be applied: Investigators seeking access to the PPMI database are asked to submit an online application then sign the data use agreement and conform with the study publications policy. Requests for accessing this database should be oriented to the PPMI Data and Publications Committee (DPC) (via: https://www.ppmi-info.org/access-data-specimens/download-data).
References
James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. The Lancet. 2018;392(10159):1789–858.
Mostafa TA, Cheng I. Parkinson’s disease detection using ensemble architecture from mr images. In: 2020 IEEE 20th international conference on bioinformatics and bioengineering (BIBE), 2020; p. 987-992. IEEE.
Pavese N, Brooks DJ. Imaging neurodegeneration in parkinson’s disease. Biochimica et Biophysica Acta (BBA)-Mol Basis Dis. 2009;1792(7):722–9.
Loane C, Politis M. Positron emission tomography neuroimaging in parkinson’s disease. Am J Transl Res. 2011;3(4):323.
Lahza H, Naveen Kumar K, Sreenivasa B, Shawly T, Alsheikhy AA, Hiremath AK, Lahza HFM. Optimization of crop recommendations using novel machine learning techniques. Sustainability. 2023;15(11):8836.
Kumar KN, Lahza H, Sreenivasa B, Shawly T, Alsheikhy AA, Arunkumar H, Nirmala C. A novel cluster analysis-based crop dataset recommendation method in precision farming. Comput Syst Sci Eng. 2023;46(3):3239–60.
Bharani B, Murtugudde G, Sreenivasa B, Verma A, Al-Yarimi FA, Khan MI, Eldin SM. Grey wolf optimization and enhanced stochastic fractal search algorithm for exoplanet detection. Eur Phys J Plus. 2023;138(5):1–11.
MS AR, Nirmala C, Aljohani M, Sreenivasa B. A novel technique for detecting sudden concept drift in healthcare data using multi-linear artificial intelligence techniques. Front Artif Intell. 2022;5: 950659.
Rahman MM, Islam MM, Manik MMH, Islam MR, Al-Rakhami MS. Machine learning approaches for tackling novel coronavirus (covid-19) pandemic. SN Comput Sci. 2021;2:1–10.
Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthcare J. 2019;6(2):94.
Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286(3):800–9.
Dong Y, Yang W, Wang J, Zhao Z, Wang S, Cui Q, Qiang Y. An improved supervoxel 3d region growing method based on pet/ct multimodal data for segmentation and reconstruction of ggns. Multimedia Tools Appl. 2020;79:2309–38.
Patil DD, Deore SG. Medical image segmentation: a review. Int J Comput Sci Mob Comput. 2013;2(1):22–7.
Ooi AZH, Embong Z, Abd Hamid AI, Zainon R, Wang SL, Ng TF, Hamzah RA, Teoh SS, Ibrahim H. Interactive blood vessel segmentation from retinal fundus image based on canny edge detector. Sensors. 2021;21(19):6380.
Alkhaldi NA, Halawani HT. Intelligent machine learning enabled retinal blood vessel segmentation and classification. Comput Mater Continua. 2023;75(1).
Teramoto A, Tsujimoto M, Inoue T, Tsukamoto T, Imaizumi K, Toyama H, Saito K, Fujita H. Automated classification of pulmonary nodules through a retrospective analysis of conventional ct and two-phase pet images in patients undergoing biopsy. Asia Ocean J Nucl Med Biol. 2019;7(1):29.
Naz SI, Shah M, Bhuiyan MIH. Automatic segmentation of pectoral muscle in mammogram images using global thresholding and weak boundary approximation. In: 2017 IEEE international WIE conference on electrical and computer engineering (WIECON-ECE), 2017. p. 199–202. IEEE.
Siri SK, Kumar SP, Latte VM. Threshold-based new segmentation model to separate the liver from ct scan images. IETE J Res. 2022;68(6):4468–75.
Khalid S, Zhu H, Yang T. Improved adaptive threshold segmentation of ultrasound medical images. In: 2021 3rd international conference on intelligent medicine and image processing, 2021. p. 1–6.
Naik MK, Panda R, Samantaray L, Abraham A. A novel threshold score based multiclass segmentation technique for brain magnetic resonance images using adaptive opposition slime mold algorithm. Int J Imaging Syst Technol. 2022;32(4):1397–413.
Nyo MT, Mebarek-Oudina F, Hlaing SS, Khan NA. Otsu’s thresholding technique for mri image brain tumor segmentation. Multimedia Tools Appl. 2022;81(30):43837–49.
Huang H, Meng F, Zhou S, Jiang F, Manogaran G. Brain image segmentation based on fcm clustering algorithm and rough set. IEEE Access. 2019;7:12386–96.
Zhang C, Shen X, Cheng H, Qian Q. Brain tumor segmentation based on hybrid clustering and morphological operations. Int J Biomed Imaging. 2019;2019.
Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S, et al. The parkinson progression marker initiative (ppmi). Prog Neurobiol. 2011;95(4):629–35.
Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The imagej ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7–8):518–29.
Senthilkumaran N, Vaithegi S. Image segmentation by using thresholding techniques for medical images. Comput Sci Eng Int J. 2016;6(1):1–13.
Ma J, Muad YA, Chen J. Visualization of medical volume data based on improved k-means clustering and segmentation rules. IEEE Access. 2021;9:100498–512.
Altini N, De Giosa G, Fragasso N, Coscia C, Sibilano E, Prencipe B, Hussain SM, Brunetti A, Buongiorno D, Guerriero A, et al. Segmentation and identification of vertebrae in ct scans using cnn, k-means clustering and k-nn. Informatics. 2021;8:40 (MDPI).
Saha PK, Udupa JK. Optimum image thresholding via class uncertainty and region homogeneity. IEEE Trans Pattern Anal Mach Intell. 2001;23(7):689–706.
Fratama RR, Partiningsih NDA, et al. Improved accuracy of vehicle counter for real-time traffic monitoring system. Transp Telecommun. 2020;21(2):125–33.
Fan Y, Rui X, Zhang G, Yu T, Xu X, Poslad S. Feature merged network for oil spill detection using sar images. Remote Sensing. 2021;13(16):3174.
Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–6.
Cao Q, Qingge L, Yang P. Performance analysis of otsu-based thresholding algorithms: a comparative study. J Sensors. 2021;2021:1–14.
Wala’a NJ, Rana J. A survey on segmentation techniques for image processing. Iraqi J Electr Electron Eng. 2021;17:73–93.
Raju PDR, Neelima G. Image segmentation by using histogram thresholding. Int J Comput Sci Eng Technol. 2012;2(1):776–9.
Popovic A, De la Fuente M, Engelhardt M, Radermacher K. Statistical validation metric for accuracy assessment in medical image segmentation. Int J Comput Assist Radiol Surg. 2007;2(3):169–81.
Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302.
Jaccard P. The distribution of the flora in the alpine zone. 1. New Phytol. 1912;11(2):37–50.
Eelbode T, Bertels J, Berman M, Vandermeulen D, Maes F, Bisschops R, Blaschko MB. Optimization for medical image segmentation: theory and practice when evaluating with dice score or jaccard index. IEEE Trans Med Imaging. 2020;39(11):3679–90.
Wahid KA, Ahmed S, He R, van Dijk LV, Teuwen J, McDonald BA, Salama V, Mohamed AS, Salzillo T, Dede C, et al. Evaluation of deep learning-based multiparametric mri oropharyngeal primary tumor auto-segmentation and investigation of input channel effects: Results from a prospective imaging registry. Clin Transl Radiat Oncol. 2022;32:6–14.
Rezatofighi H, Tsoi N, Gwak J, Sadeghian A, Reid I, Savarese S. Generalized intersection over union: a metric and a loss for bounding box regression. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2019; p. 658–666.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Not applicable.
Conflict of Interest
The authors declare that they have no conflict of interest.
Authors’ contributions
All the authors contributed to the study conception, design, and paper selection. The first draft of the paper was written by ZM and all the authors commented on previous versions of the article. FBR and KN revised the work, and have approved the submitted version.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Soft Computing in Engineering Applications” guest edited by Kanubhai K. Patel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Maalej, Z., Ben Rejab, F. & Nouira, K. Comparative Study of Segmentation Techniques for Basal Ganglia Detection Based on Positron Emission Tomography Images. SN COMPUT. SCI. 5, 364 (2024). https://doi.org/10.1007/s42979-024-02677-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02677-9