Abstract
Biosurfactants are one of the microbial bioproducts that are in most demand from microbial-enhanced oil recovery (MEOR). We isolated and screened potential biosurfactant-producing bacteria, followed by biosurfactant production and characterization, and a simulation of the MEOR application to biosurfactants in a sand-packed column. Isolate screening was conducted based on qualitative (hemolytic blood assay and oil-spreading test) and semi-qualitative (emulsification assay and interfacial tension measurement) parameters. Bacterial identification was performed using 16S rRNA phylogenetic analysis. Sequential isolation yielded 32 bacterial isolates, where Pseudomonas sp. G3 was able to produce the most biosurfactant. Pseudomonas sp. G3 had the highest emulsification activity (Ei = 72.90%) in light crude oil and could reduce the interfacial tension between oil and water from 12.6 to 9.7 dyne/cm with an effective critical-micelle concentration of 0.73 g/L. The Fourier transform infrared spectrum revealed that the biosurfactant produced was a glycolipid compound. A stable emulsion of crude extract and biosurfactant formed at pH 2–12, up to 100 °C, and with a NaCl concentration of up to 10% (w/v) in the response-surface method, based on the Box–Behnken design model. The sand-packed column experiment with biosurfactant resulted in 20% additional oil recovery. Therefore, this bacterium and its biosurfactant show potential and the bacterium is suitable for use in MEOR applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The demand for energy resources continues to increase with time. Developments in renewable energy are expected to provide sustainable energy and environmentally friendly industries. However, many projects that are related to renewable energy face challenges, including technical, social, and economic challenges. Existing energy resources should optimize production, while avoiding critical environmental risks. Microbial-enhanced oil recovery (MEOR) is an alternative approach to optimize oil production from existing reservoirs. This method is considerably more economic and environmentally friendly than other EOR methods. Energy that is used in microbial processes to enhance oil recovery does not depend on the price of crude oil. Microbes can growth independently under many conditions and produce large amounts of useful products rapidly from cheap, renewable materials that are available in large quantities. As a biological agent, microbial bioproducts are often biodegradable, which results in lower levels of pollution and a low toxicity (Youssef et al. 2009; Khire 2010). MEOR uses microbial activities and various bioproducts to help release residual oil that is trapped inside the rock pores and to stimulate oil flow to the production wells (Safdel et al. 2017).
Microorganisms produce biosurfactants, and the latter are important in the MEOR mechanism. Biosurfactants act as surface-active molecules that reduce the interfacial tension (IFT) between different fluid components, and enhance pseudosolubilization of oil in water by creating smaller oil droplets (Khire 2010). Injection of partially purified biosurfactants has increased the amount of recovered oil to 40% (Mcinerney et al. 2003). Compared with chemically synthesized surfactants, biosurfactants are biodegradable, non-toxic, characteristically diverse, and stable under extreme conditions. Biosurfactant production can be significantly more affordable, as it may be produced using biomass waste (Gautam and Tyagi 2008; Jing et al. 2011; Dhasayan et al. 2014).
Limited studies exist on the in situ production of biosurfactants for MEOR implementation and biosurfactant stability under various physiological conditions. This study aimed to isolate, screen, and identify potential biosurfactant-producing bacteria from oil reservoir samples. We characterized the biosurfactant stability under several physiological conditions using Fourier transform infrared (FT-IR) spectroscopy and response-surface methodology. MEOR simulations were performed on a sand-packed column to determine the biosurfactant’s applicability to recover residual crude oil.
Materials and methods
Material
Samples were from a petroleum reservoir in South Sumatra. Sample that contained crude oil (heavy oil) and formation water (brine) were extracted from its wellhead. All the samples were kept in a cold box during transportation and they were kept at 4 °C in the laboratory until they were analyzed. Light crude oil was utilized in these experiments. The characteristics of the crude oil and formation brine are listed in Tables 1 and 2.
Isolation of biosurfactant-producing bacteria
Isolation was carried out in two sequential stages (Halim et al. 2008) to obtain bacteria with varying oil-degrading potential. Sample was added into Stone Mineral Salt Solution (SMSS) medium with 0.1% (w/v) yeast extract and incubated at 50 °C and 70 °C with 120 rpm agitation for 7 days. A plate count was conducted every 48 h using the pour method on Nutrient Agar Difco™. Residual oil from the first stage was used as a substrate in the second stage of sequential isolation using the same procedure. Purification of potential biosurfactant-producing bacteria was conducted with a four-way streak method on NA medium. The purified cultures were used for further screening and analysis.
Screening of potential biosurfactant-producing bacteria
Screening was performed based on two qualitative (hemolytic blood assay and oil-spreading test) and two semi-qualitative (emulsification index and IFT measurement) methods. Preliminary screening used the hemolytic blood assay method according to Youssef et al. (2004). Further confirmation of isolate biosurfactant production was done using the oil-spreading test, based on a study by Satpute et al. (2010). The emulsification activity of the produced biosurfactant to emulsify crude oil and water was investigated from the emulsification index (Ei/E24) (Satpute et al. 2010). IFT measurements were performed using a Surface Tensiomat Model 21 (Fisher Scientific™, USA) with a 6-cm ring diameter to investigate the ability of the biosurfactant-producing bacteria to decrease the IFT between the oil and the water.
Critical-micelle-concentration determination
The critical-micelle concentration (CMC) was obtained by measuring the IFT and the biosurfactant concentration (g/L) at a certain time. The effective CMC was determined when an increase in the biosurfactant concentration did not result in a significant decrease in IFT (Satpute et al. 2010).
Identification of potential biosurfactant-producing bacteria
The selected biosurfactant-producing bacteria was identified using a 16S ribosomal DNA analysis and was designated MEOR_G3. DNA sequencing was performed using a Macrogen™ Korea with 785F and 907R universal primers. All the base sequences that were generated from sequencing were cross-checked manually using Bioedit and similarities were determined using BLASTN and RDP. Multiple sequence alignment was conducted using CLUSTAL X, and a phylogenetic tree was constructed using MEGA6 Software.
Extraction of biosurfactant
The growth and biosurfactant production profile of the selected bacterium (MEOR_G3) were studied using the total-plate-count method. Batch production of the biosurfactant was performed in SMSS medium with a 2% sample and 0.1% yeast extract with 120 rpm agitation at 50 °C. Biosurfactant extraction was carried out according to Smyth et al. (2010).
Characterization of biosurfactant by Fourier transform infrared
A crude biosurfactant extract was dried to powder and characterized using Fourier transform infrared (FT-IR) to elucidate its functional groups. Scanning was conducted from 4000 to 400/cm (Sriram et al. 2011). The sample was prepared by grinding, and its formation into a pellet with KBr. The output from the scanning process was the transmittance percentage in the infrared spectra.
Biosurfactant-stability test
The Box–Behnken design model was used to study the interactive effects of the biosurfactant emulsification stability at different pH conditions, temperatures, and salinities. The temperature ranged from 40 to 120 °C, the pH from 2 to 12, and the salinity from 2 to 10% (w/v) NaCl. The pH was adjusted using 1 N NaOH and 1 N HCl. The stability of a formed emulsion was determined by measuring the Ei as a generated response after 24 h of incubation. All the experiments were performed in triplicate. A regression equation was derived from the response analysis, including the determinant coefficients (F and R2), which were evaluated to generate contour plots. These plots were used to determine the interaction between different factors and to predict the emulsification stability at certain points (Dhasayan et al. 2014). All the statistical analyses were carried out using Minitab™ 17 Software.
Sand-packed column assay
Biosurfactant application for enhanced oil recovery was evaluated using a modified sand-packed column described by Suthar et al. (2008). Glass columns packed with sand (50 mesh particle size) were first saturated with brine. The pore volume (PV, ml) of the sand-packed column was calculated by measuring the volume of brine required to saturate the column. The porosity (%) of the column was calculated as the PV divided by the total volume of the empty column. In the second step, light crude oil was flooded into the column until no more brine emerged from the base of the column. The original oil in place (OOIP) was calculated as the volume of crude oil retained in the column. The initial oil saturation (Soi, %) (Eq. 1) and initial brine saturation (Swi, %) (Eq. 2) were calculated as follows:
The column was flooded again with brine to obtain the residual oil saturation (Sor, %). The amount of oil recovered, which is the so-called oil recovered after brine flooding (Sorwf, ml), was used to calculate the residual oil saturation (Sor) (Eq. 3) as follows:
The residual oil in the column was flooded with a 0.6 pore volume of crude biosurfactant and incubated at 50 °C for 24 h. The column was flooded again with brine and the amount of oil recovered using crude biosurfactant (oil recovered after biosurfactant flooding (Sorbf, ml)) was used to calculate the additional oil recovery (AOR, %) (Eq. 4) as follows:
The control column was flooded with brine only and was incubated under the same conditions.
Results and discussion
Sequential isolation of microorganisms and screening results
The principal of sequential isolation used in this study was the pattern of hydrocarbon susceptibility to degradation by bacteria from the easiest to the hardest chain, which can be written as: alkane > light aromatic > cycloalkane > heavy aromatic > asphalt (Van Hamme et al. 2003). Sequential isolation was expected to capture diverse degrading capabilities of indigenous bacteria for different oil fractions (Halim et al. 2008). The first stage would isolate bacteria that can degrade light-oil fractions, whereas the second stage would isolate bacteria that metabolized a heavier oil fraction as substrate. Bacterial activities on the heavier fraction of crude oil degradation will decrease the oil viscosity, and thus, have a high potential to be used in MEOR (Purwasena et al. 2014).
Sequential isolation resulted in 18 bacterial isolates from stage one and 14 isolates from stage two (Table 3). Gram staining showed that 30 isolates were Gram-negative rods and two isolates were Gram-positive cocci. Some Gram-negative bacteria dominate the subsurface and oil-contaminated environment because they have adapted to live in a hydrocarbon mixture, such as poly- and mono-aromatics, as reported in several studies (Batista et al. 2006; Lazaroaie 2010; Shoeb et al. 2015). Several possible mechanisms of this physiological adaptation are the impermeability of cell membranes to toxic hydrocarbon, the formation of heat-stable proteins as a part of the hydrocarbon efflux pumps, and the DNA-repairing ability (Madigan et al. 2013).
All isolated bacteria were screened for a high biosurfactant-producing activity. Screening was carried out against qualitative (hemolytic blood assay and oil spreading test) and semi-quantitative (emulsification index) parameters. Based on the result, an isolate that was coded G3 showed the highest emulsification activity (Ei = 72.90%). This result indicates that isolate G3 was a biosurfactant-producing bacterium with the highest MEOR potential among other isolates. This bacterium could grow at a moderately high temperature of up to 55 °C. The negative correlation between its hemolytic type (G3 is of gamma type) and its emulsification activity may result because the biosurfactant failed to diffuse to the surrounding blood agar media, as reported and reviewed in several studies (Marchant and Banat 2014; Walter et al. 2010). This prevents the biosurfactant from exhibiting hemolytic activity.
Biosurfactant ability to reduce IFT between oil and water
Biosurfactant that is produced by bacteria in the MEOR mechanism may increase the amount of recovered oil by reducing the IFT between oil and water inside the reservoirs, which modifies the pore-rock wettability by changing the rock properties from oil-wet to water-wet, and by emulsifying hydrocarbons (Perfumo et al. 2010). These surface-active molecules can disperse oil components in water by establishing a bridge between fluids with different polarities, such as oil–water, by reducing free energy per unit area by forming micelles that are surrounded the oil droplets, and by increasing the oil pseudosolubilization in water (Khire 2010).
The G3 bacterium could decrease the IFT between oil and water from 12.9 to 9.7 dyne/cm (Fig. 1). At 0.73 g/L, a maximum reduction of IFT was achieved (from 12.6 to 11.3 dyne/cm), which indicates that the effective CMC of the biosurfactant that was produced by the G3 was 0.73 g/L. A slight difference between the reported CMC value in this experiment and previous studies may result from some inherent properties of the biosurfactant, such as the hydrophobic chain length, purity, growth-media composition, and strain of producing bacteria (Abdel-Mawgoud et al. 2009). From the CMC determination, a G3 biosurfactant was observed to exhibit good tension-active properties alongside its high emulsification activity. Hence, this molecule has potential for application in MEOR.
Phylogenetic analysis and identification using 16S rRNA sequences
In Nutrient Agar media, G3 appeared to be yellow to pale yellow, and microscopic observation revealed that the Gram-negative stained cells were rod-shaped. The colony could grow up to 55 °C. The sequencing of ribosomal DNA coded by the 16S rRNA gene acquired almost 99% of the total length of the 16S rRNA sequences (1484 bp from ± 1500 bp). Early analysis was by RDPII and NCBI BlastN and G3 was classified as Gammaproteobacteria within the Xanthomonadaceae family and Pseudoxanthomonas genus. BlastN confirmed G3’s 99% identity (e value 0.00) with Pseudoxanthomonas taiwanensis. Further analysis using phylogenetic tree reconstruction as depicted in Fig. 2 revealed that G3 was included in the cluster group of the Pseudoxanthomonas taiwanensis strain NBRC 101072, P. taiwanensis CB-226, and P. suwonensis strain 4M1.
Previous studies reported the isolation of Pseudoxanthomonas sp. from global hydrocarbon ecosystems, such as from oil sludge, BTEX-contaminating soil, and mining coal (Lee et al. 2008; Harada et al. 2006; Kumari et al. 2011; Young et al. 2007); Nayak et al. (2009) reported the successful isolation of Pseudoxanthomonas sp. PNK-04, which could produce rhamnolipid biosurfactant from coal samples. Chang et al. (2005) were able to isolate Pseudoxanthomonas kaohsiungensis from oil sludge samples in Taiwan with a high emulsification activity on crude oil and they were able to lower the surface tension from 68 to 32.6 dyne/cm.
Finally, in accordance with the 16S rRNA sequences and phylogenetic analysis, the G3 bacterium was named Pseudoxanthomonas sp. G3.
Growth curve and biosurfactant production of Pseudoxanthomonas sp. G3
To investigate the pattern of biosurfactant synthesis by Pseudoxanthomonas sp. G3, the bacterium growth was studied in batch mode using SMSS media with heavy oil as the main carbon source, in parallel with biosurfactant production measurements. The kinetics of biosurfactant production is important for larger industrial-scale development and practical purposes, such as MEOR. The highest cost spent on biosurfactant production is on downstream processing, e.g., extraction and purification. Thus, biosurfactants with a faster production time and fewer impurities will be very promising for industry (Mulligan et al. 2014).
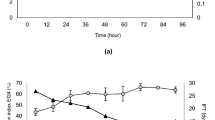
Figure 3 shows that a lag phase appeared in the first 12 h, then the bacterium entered an exponential phase in the next 12 h with a constant and maximum growth rate (0.434 CFU/h). After 24 h, the cells reached a stationary phase. In parallel with the growth phase, biosurfactant production by the bacterium was low during the initial 12 h, increased significantly within the next 12 h until 48 h of incubation, and reached a maximal concentration at a late-exponential-early stationary phase. Therefore, it can be concluded that biosurfactant that is produced by the bacterium can be considered to be a growth-associated metabolite. The result concurs with the previous report by Nayak et al. (2009).
In the late-exponential-early stationary phase, maximum biosurfactant production resulted because of mineral limitations in media, such as N, Ca, Mg, and P, which had been exhausted during the logarithmic-growth periods (Mulligan and Gibbs 1989 in Mulligan et al. 2014). The decrease in biosurfactant concentration as the cell entered the late-exponential phase to the early death phase may be caused by biosurfactant interaction with the cell membrane, such as a phospholipid, which has a similar structure (Hommel 1990). As a result, the bacterium may have experienced an excessive toxic effect. Consequently, the biosurfactant concentration in the medium declined at the end of the incubation period.
FT-IR analysis of biosurfactant molecule
Preliminary analysis to characterize the biosurfactant produced by Pseudoxanthomonas sp. G3 was conducted using the qualitative blue agar plate method as described by Siegmund and Wagner (1991). The bacterial colony produced a dark blue color on the media, which suggests the production of a glycolipid class of biosurfactants by the bacterium. Further characterization was conducted using FT-IR spectroscopy.
The IR spectra (Fig. 4) showed a maximum absorption at several wavenumber locations, which was caused by the presence of distinct functional groups (Coates 2016). The most important peaks at 3388–3136/cm (stretching of O–H and C–H bonds) and several wavenumber positions at 1647, 1543, and 1386/cm correspond to C=O stretching (part of carbonyl functional group), C–C stretching, and α-CH2 and α-CH3 bending (representing aliphatic chains), respectively. In the fingerprint regions (1500–500/cm), the absorption bands at 1103, 964, 829, and 609/cm confirmed the presence of C–O bonds (which indicates part of the hydrophilic regions in glycoside moieties), bending of trans C–H and C–H bonds, C–C vibrations, and bending of O–H bonds, respectively.
According to IR spectral analysis, biosurfactant produced by Pseudoxanthomonas sp. G3 was a glycolipid. The structure and functional groups in the molecule were similar to the rhamnolipid and glycolipid structure with the presence of –OH, CO, –COOH, CH2, and CH3 groups in the structure (Nitschke et al. 2011).
Effects of pH, temperature, and salinity on biosurfactant stability
The effectiveness of biosurfactant produced by Pseudoxanthomonas sp. G3 for MEOR implementation was evaluated by studying its emulsification stability that was influenced by the interactive effect of pH, temperature, and salinity. The physiological factors were considered to be the most critical environmental parameter in the MEOR mechanisms, specifically for biosurfactant injection, as reported by Al-Sulaimani et al. (2011). Biosurfactant must be stable for 50–80 °C, a wide range of pH values, and various salinity concentrations that simulate oil reservoir conditions. A good biosurfactant emulsifying property is its potential to instigate oil–water emulsification (Perfumo et al. 2010).
To determine the relationship caused by the interactive effect of previous critical factors with biosurfactant stability, polynomial regression equation was constructed using the Box–Behnken model of the response-surface methodology. The measured response was the emulsification index (Ei) after 24 h with a 15-min temperature exposure. ANOVA statistical analysis from the generated response in term of Ei (Table 4) showed that the most significant interactive effect of the environmental factor on biosurfactant emulsifying activity was the interaction between pH and temperature (p < 0.05). The analysis produced the following equation for Ei calculation (Eq. 5):
The cell-free supernatant that contained biosurfactant produced by Pseudoxanthomonas sp. G3 could maintain a high emulsification activity up to 50% in a wide pH range (2–12) and at a NaCl concentration of up to 10% (w/v). The biosurfactant was relatively stable up to 100 °C. The ability to preserve a moderate salinity environment implies that the biosurfactant was halophilic (Chandankere et al. 2013) and the moderately strong ionic tolerance indicated its suitability for MEOR under high-salinity conditions (Shavandi et al. 2011). Several previous studies have reported that glycolipid biosurfactant could maintain its activity in a pH range of 2–10, at a moderate NaCl concentration, and at extreme temperatures (Mnif and Ghribi 2015). The contour plot (Fig. 5) depicts the interactive effect of pH, salinity, and temperature on the emulsion stability of a crude-form biosurfactant by the selected bacterium. The unique tolerance of biosurfactant produced by Pseudoxanthomonas sp. G3 makes this bacterium a good candidate for MEOR application in a wide range of pH, salinities, and temperatures, including for bioproduct injection.
Sand-packed column experiment
To evaluate the applicability of crude biosurfactant produced by Pseudoxanthomonas sp. G3 for ex situ MEOR, biosurfactant was flooded into the sand-packed column experiment to release residual oil in the column. We found a different structure and permeability zone from sand packed in the column as reflected from the variation in PV and all other parameters, including the amount of residual oil in the column. Biosurfactant flooding in two sand-packed columns yielded 38.46–42.11% oil recovery. Compared with the control, biosurfactant flooding increased the oil recovery by 20% (18.4–42.11%). A summary of the oil recovery in the sand-packed column using brine flooding and a crude biosurfactant of Pseudoxanthomonas sp. G3 is shown in Table 5. Previously, a sand-packed column experiment that used crude biosurfactant flooding produced by different species of Bacillus spp. and Fusarium sp. BS-8 resulted in 24.5–46% additional oil recovery (Suthar et al. 2008 and; Qazi et al. 2013). Gudina et al. (2013) and Sun et al. (2018) reported that for the MEOR experiment, the oil recovery from OOIP was below 50%.
Conclusions
A thermophilic biosurfactant-producing microorganism was isolated from a heavy oil reservoir and identified as Pseudoxanthomonas sp. G3. The potential use of the biosurfactant produced by this bacterium for MEOR lies in its high emulsification activity in light crude oil (Ei value 72.90%) and its ability to lower the IFT from 12.9 to 9.7 dyne/cm with an effective CMC of 0.73 g/l. The FT-IR spectra show that the produced biosurfactant was glycolipid in nature. The crude extract of the produced biosurfactant was stable at high temperatures and in a moderately saline environment, and could maintain its activity over a wide range of pH values based on the Box–Behnken model design. A sand-packed column experiment with the biosurfactant resulted in 20% additional oil recovery. Therefore, this bacterium and its biosurfactant could be suitable for use in MEOR.
References
Abdel-Mawgoud A, Aboulwafa W, Hassouna N (2009) Characterization of rhamnolipid produced by Pseudomonas aeruginosa isolate Bs20. Appl Biochem Biotechnol 15:329–345
Al-Sulaimani H, Al-Wahaibi Y, Al-Bahry S, Elshafie A, Al-Bermani A, Joshi S, Zargari S (2011) optimization and partial characterization of biosurfactants produced by bacillus species and their potential for ex-situ enhanced oil recovery. SPE J 16(03):672–682
Batista SB, Mounteer AH, Amorim FR, Totola MR (2006) Isolation and characterization of biosurfactant/bioemulsifier-producing bacteria from petroleum contaminated sites. Biores Tech 97:868–875
Chandankere R, Jun Y, Choi MMF, Masakorala K, Chan Y (2013) An efficient biosurfactant-producing and crude-oil emulsifying bacterium Bacillus methylotrophicus USTBa isolated from petroleum reservoir. Biochem Eng J 74:46:53
Chang JS, Chou CL, Lin GH, Sheu SY, Chen YM (2005) Pseudoxanthomonas kaohsiungensis, sp. nov., a novel bacterium isolated from oil-polluted site produces extracellular surface activity. Syst Appl Microbiol 28(2):137–144
Coates J (2016) Interpretation of infrared spectra, a practical approach. In: Meyers RA (ed) Encyclopedia of Analytical Chemistry. Wiley, USA, pp 10815–10837
Dhasayan A, Kiran GS, Selvin J (2014) Production and characterisation of glycolipid biosurfactant by Halomonas sp. MB-30 for potential application in enhanced oil recovery. J Appl Biochem Biotechnol 174(7):2571–2584
Gautam KK, Tyagi VK (2006) Microbial surfactants: a review. J Oleo Sci 55(4):155–166
Gudina EJ, Pereira JFB, Costa R, Coutinho JAP, Teixeira JA, Rodrigues LR (2013) Biosurfactant-producing and oil-degrading Bacillus subtilis strains enhanced oil recovery in laboratory sand-pack columns. J Hazard Mater 261:106–113
Halim AY, Andromeda A, Mustafa I, Haro M, Purwasena IA, Juli N, Siregar S (2008) The laboratory study of indigenous bacterial activities on physical oil sample properties from west java oil reservoir: implication for MEOR (Microbial Enhanced Oil Recovery) application. Professional Paper of 10th National Symposium and Congress, Ikatan Ahli Teknik Perminyakan Indonesia (IATMI)
Harada RM, Campbell S, Li QX (2006) Pseudoxanthomonas kalamensis sp. nov., a novel gammaproteobacterium isolated from Jonhston Atoll, North Pacific Ocean. J Syst Evol Microbiol 56:1103–1107
Hommel RK (1990) Formation and physiological role of biosurfactants produced by hydrocarbon-utilizing microorganisms. Biodegradation 1(2):107–119
Jing W, Guang J, Jing T, Hongdan Z, Hanping D, Li Y (2011) Functional characterization of a biosurfactant-producing thermo-tolerant bacteria isolated from an oil reservoir. Pet Sci 8:353–356
Khire JM (2010) Bacterial biosurfactants, and their role in microbial enhanced oil recovery (MEOR). In: Ramkrishna S (ed) Biosurfactants. Landes Bioscience and Springer Science + Business Media, New York, pp 146–157
Kumari K, Sharma P, Tyagi K, Lal R (2011) Pseudoxanthomonas indica sp. nov., isolated from a hexachlorocyclohexane dumpsite. J Syst Evol Microbiol 61:2107–2111
Lazaroaie MM (2010) multiple responses of gram-positive and gram-negative bacteria to mixture of hydrocarbons. Braz J Microbiol 41:649–667
Lee DS, Ryu SH, Hwang HW, Kim YJ, Park M, Lee JR, Lee SS, Jeon CO (2008) Pseudoxanthomonas sacheonensis sp. nov., isolated from BTEX contaminated soil in Korea, transfer of Stenotrophomonas dokdonensis Yoon et al. 2006 to the genus Pseudoxantho monas as Pseudoxanthomonas dokdonensis comb. nov. and emended description of the genus Pseudoxanthomonas. J Syst Evol Microbiol 58:2235–2240
Madigan MT, Martinko JM, Stahl DA, Clark DP (2013) Brock biology of microorganisms, 13th edn. Pearson International, New York
Marchant R, Banat IM (2014) Protocols for measuring biosurfactant production in microbial cultures hydrocarbon and lipid microbiology protocols. In: McGenity TJ et al (eds) Hydrocarbon and lipid microbiology protocols. Springer protocols handbooks. Springer Verlag, Berlin, pp 119–128
Mcinerney MJ, Han SO, Maudgalya S, Mouttaki H, Folmsbee M, Knapp R, Nagle D, Jackson BE, Stuadt M, Frey W (2003) Development of more effective biosurfactants for enhanced oil recovery. (No. DOE/BC/15113-2). National Petroleum Technology Office, U.S. Department of Energy. Tulsa, Oklahoma, USA
Mnif I, Ghribi D (2015) Microbial derived surface active compounds: properties and screening concept. World J Microbiol Biotechnol 31(7):1001–1020
Mulligan CN, Gibbs BF (1989) Correlation of nitrogen metabolism with biosurfactant production by Pseudomonas aeruginosa. Appl Environ Microbiol 55(11):3016–3019
Mulligan CN, Sharma SK, Mudhoo A (2014) Biosurfactants: research trends and applications. CRC Press, Florida
Nayak AS, Vijaykumar MH, Karegoudar TB (2009) Characterization of biosurfactant produced by Pseudoxanthomonas sp. PNK-04 and its application in bioremediation. Intern Biodet Biodeg 63:73–79
Nitschke M, Costa SGVAO, Contiero J (2011) Rhamnolipids and PHAs: recent reports on Pseudomonas-derived molecules of increasing industrial interest. Proc Biochem 46:621–630
Perfumo A, Rancich I, Banat IM (2010) Possibilities and challenges for biosurfactants use in petroleum industry. In: Sen R (ed) Biosurfactants. Landes, (ed) Bioscience and Springer Science + Business., Media, New York. Springer, New York, pp 135–145
Purwasena IA, Sugai Y, Sasaki K (2014) Estimation of the potential of an anaerobic thermophilic oil-degrading bacterium as a candidate for MEOR. J Petrol Explor Prod Technol 4(2):189–200
Qazi MA, Subhan M, Fatima N, Ali MI, Ahmed S (2013) Role of biosurfactant produced by Fusarium sp. BS-8 in enhanced oil recovery (EOR) through sand pack column. Int J Biosci Bioinforma 3(6):598
Safdel M, Anbaz MA, Daryasafar A, Jamialahmadi M (2017) Microbial enhanced oil recovery, a critical review on worldwide implemented field trials in different countries. Renew Sustain Energy Rev 74:159–172
Satpute SK, Banpurkar AG, Dhakephalkar PK, Banat IM, Chopade BA (2010) Methods for investigating biosurfactants and bioemulsifiers: a review. Crit Rev Biotechnol 30:127–144
Shavandi M, Mohebali G, Haddadi A, Shakarami H, Nuhi A (2011) Emulsification potential of a newly isolated biosurfactant-producing bacterium, Rhodococcus sp. strain TA6. Colloids Surface B Biointerfaces 82:477–482
Shoeb E, Ahmed N, Akhter J, Ansari M (2015) Screening and characterization of biosurfactant-producing bacteria isolated from the Arabian Sea coast of Karachi. Turk J Biol 39(2):210–216
Siegmund I, Wagner F (1991) New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotech Tech 5(4):265–268
Smyth TJP, Perfumo A, McClean S, Marchant R, Banat IM (2010) Isolation and analysis of lipopeptides and high molecular weight biosurfactants. In: Timmis KN (ed) handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Berlin, pp 3687–3704
Sriram MI, Kalishwaralal K, Deepak V, Gracerosepat R, Srisakhti K, Gurunathan S (2011) Biofilm inhibition and antimicrobial action of lipopeptide biosurfactant produced by heavy metal tolerant strain Bacillus cereus NK1. Colloids Surf B 85:174–181
Sun G, Hu J, Wang Z, Li X, Wang W (2018) Dynamic investigation of microbial activity in microbial enhanced oil recovery (MEOR). Petrol Sci Technol 36(16):1–7
Suthar H, Hingurao K, Desai A, Nerurkar A (2008) Evaluation of bioemulsifier mediated microbial enhanced oil recovery using sand pack column. J Microbiol Meth 75(2):225–230
Van Hamme JD, Singh A, Ward OP (2003) Recent advances in petroleum microbiology. Microb Mol Biol 67:503–549
Walter V, Syldatk C, Hausmann R (2010) screening concepts for the isolation biosurfactant producing microorganisms in biosurfactants. In: R Sen (ed) Biosurfactants. Landes Bioscience and Springer Science + Business Media, New York, pp 1–13
Young CC, Ho M, Arun AB, Chen W, Lai W, Shen F, Rekha PD, Yassin AF (2007) Pseudoxanthomonas spadix sp. nov., isolated from oil-contaminated soil. J Syst Evol Microbiol 57:1823–1827
Youssef NH, Duncan KE, Nagle DP, Savage KN, Knapp RM, Mclnerney MJ (2004) Comparison of methods to detect biosurfactant production by diverse microorganism. J Microbiol Meth 56(3):339–347
Youssef N, Elshahed MS, McInerney MJ (2009) Microbial processes in oil fields: culprits, problems, and opportunities. Adv Appl Microbiol 66:141–251
Acknowledgements
The authors thank Oil and Gas Recovery for Indonesia (OGRINDO) ITB, Pertamina-EP for sample preparation, The Indonesia Ministry of Education Directorate General of Higher Education, DIKTI, for facilitating and funding the international research collaboration scheme, and Japan Student Service Association (JASSO) for providing a scholarship. We thank Qonita Afinanisa from Institut Teknologi Bandung and Laura Kuhar, PhD, from Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Astuti, D.I., Purwasena, I.A., Putri, R.E. et al. Screening and characterization of biosurfactant produced by Pseudoxanthomonas sp. G3 and its applicability for enhanced oil recovery. J Petrol Explor Prod Technol 9, 2279–2289 (2019). https://doi.org/10.1007/s13202-019-0619-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13202-019-0619-8