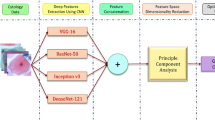
Abstract
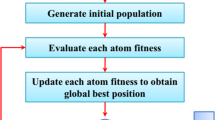
Over 500 K (per year) cervical cancer cases are reported with a high mortality rate (6–9%). Automatically detecting cervical cancer using the Computer-Aided Diagnosis (CAD) tool at an early stage is important since it leads to successful treatment as pathologists. In this paper, we propose a tool that classifies cervical cancer cases from Pap smear cytology images using deep features. The proposed tool constitutes a Convolutional Neural Network (CNN) and a metaheuristic evolutionary algorithm called Opposition-based Harmony Search Algorithm (O-bHSA) for deep feature section. These features are classified using standard classifiers: SVM, MLP, and KNN. On two different publicly available datasets: Pap smear and liquid-based cytology, the proposed tool outperforms not only seven well-known optimization algorithms but also state-of-the-art methods. Codes are publicly available on GitHub.





Similar content being viewed by others
Data availability
In the present work, we have used two datasets, (a) Mendeley Liquid-Based Cytology (MLBC) [34] and (b) SIPaKMeD Pap Smear dataset [16]. The data sets are publicly availble at “https://data.mendeley.com/datasets/zddtpgzv63/4” and “https://www.cs.uoi.gr/~marina/sipakmed.html” respectively.
References
Mitra S, Das N, Dey S, Chakraborty S, Nasipuri M, Naskar MK (2022) Cytology image analysis techniques toward automation. ACM Comput Surv 54(3):1–41
Santosh KC, Das N, Ghosh S (2022) Chapter 4—cytology image analysis. In: Santosh KC, Das N, Ghosh S (eds) Deep learning models for medical imaging. Primers in biomedical imaging devices and systems. Academic Press, Cambridge, pp 99–123
Rahaman MdM, Li C, Wu X, Yao Y, Hu Z, Jiang T, Li X, Qi S (2020) A survey for cervical cytopathology image analysis using deep learning. IEEE Access 8:61687–61710
Dholey M, Sarkar A, Maity M, Giri A, Sadhu A, Chaudhury K, Das S, Chatterjee J (2018) A computer vision approach for lung cancer classification using fnac-based cytological images. In: Proceedings of 2nd international conference on computer vision and image processing. Springer, Berlin, pp 181–195
Allehaibi KHS, Nugroho LE, Lazuardi L, Prabuwono AS, Mantoro T et al (2019) Segmentation and classification of cervical cells using deep learning. IEEE Access 7:116925–116941
Alyafeai Z, Ghouti L (2020) A fully-automated deep learning pipeline for cervical cancer classification. Expert Syst Appl 141:112951
Kuko M, Pourhomayoun M (2019) An ensemble machine learning method for single and clustered cervical cell classification. In: 2019 IEEE 20th international conference on information reuse and integration for data science (IRI). IEEE, USA, pp 216–222. https://doi.org/10.1109/IRI.2019.00043
Sen A, Mitra S, Chakraborty S, Mondal D, Santosh KC, Das N (2022) Ensemble framework for unsupervised cervical cell segmentation. In: 2022 IEEE 35th international symposium on computer-based medical systems (CBMS). IEEE, USA, pp 345–350
Iliyasu AM, Fatichah C (2017) A quantum hybrid PSO combined with fuzzy k-NN approach to feature selection and cell classification in cervical cancer detection. Sensors 17(12):2935
Tseng C-J, Lu C-J, Chang C-C, Chen G-D (2014) Application of machine learning to predict the recurrence-proneness for cervical cancer. Neural Comput Appl 24(6):1311–1316
William W, Ware A, Basaza-Ejiri AH, Obungoloch J (2018) A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images. Comput Methods Programs Biomed 164:15–22
Nanni L, Ghidoni S, Brahnam S (2017) Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recognit 71:158–172
Zare MR, Alebiosu DO, Lee SL (2018) Comparison of handcrafted features and deep learning in classification of medical x-ray images. In: 2018 Fourth international conference on information retrieval and knowledge management (CAMP). IEEE, USA, pp 1–5. https://doi.org/10.1109/INFRKM.2018.8464688
Kwon M (2018) Multi-label Classification of Single and Clustered Cervical Cells Using Deep Convolutional Networks. California State University, Los Angeles
Phoulady HA, Mouton PR (2018) A new cervical cytology dataset for nucleus detection and image classification (cervix93) and methods for cervical nucleus detection. arXiv preprint. abs/1811.09651. arXiv:1811.09651
Plissiti ME, Dimitrakopoulos P, Sfikas G, Nikou C, Krikoni O, Charchanti A (2018) SIPAKMED: a new dataset for feature and image based classification of normal and pathological cervical cells in pap smear images. In: 2018 25th IEEE international conference on image processing (ICIP). IEEE, USA, pp 3144–3148. https://doi.org/10.1109/ICIP.2018.8451588
Wu M, Yan C, Liu H, Liu Q, Yin Y (2018) Automatic classification of cervical cancer from cytological images by using convolutional neural network. Biosci Rep 28; 38(6):BSR20181769. https://doi.org/10.1042/BSR20181769
Basak H, Kundu R, Chakraborty S, Das N (2021) Cervical cytology classification using PCA and GWO enhanced deep features selection. SN Comput Sci 2(5):369. https://doi.org/10.1007/s42979-021-00741-2
Sompawong N, Mopan J, Pooprasert P, Himakhun W, Suwannarurk K, Ngamvirojcharoen J, Vachiramon T, Tantibundhit C (2019) Automated pap smear cervical cancer screening using deep learning. In: 2019 41st Annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, USA, pp 7044–7048. https://doi.org/10.1109/EMBC.2019.8856369
Meiquan X, Weixiu Z, Yanhua S, Junhui W, Tingting W, Yajie Y, Meng Z, Zeji Z, Longsen C (2018) Cervical cytology intelligent diagnosis based on object detection technology. In: 1st Conference on medical imaging with deep learning (MIDL 2018), Amsterdam, The Netherlands, 7th January 2018, pp 1–9
Li C, Xue D, Zhou X, Zhang J, Zhang H, Yao Y, Kong F, Zhang L, Sun H (2019) Transfer learning based classification of cervical cancer immunohistochemistry images. In: Proceedings of the third international symposium on image computing and digital medicine. Association for Computing Machinery New York, NY, United States, pp 102–106
Win KP, Kitjaidure Y, Hamamoto K, Aung TM (2020) Computer-assisted screening for cervical cancer using digital image processing of pap smear images. Appl Sci 10(5):1800
Mitra S, Dey S, Das N, Chakrabarty S, Nasipuri M, Naskar MK (2020) Identification of malignancy from cytological images based on superpixel and convolutional neural networks. Springer Singapore, Singapore, pp 103–122
Geem ZW, Kim JH, Loganathan GV (2001) A new heuristic optimization algorithm: harmony search. Simulation 76(2):60–68
Mitchell M (1996) An introduction to genetic algorithms. MIT Press, Cambridge
Singh RP, Mukherjee V, Ghoshal SP (2013) The opposition-based harmony search algorithm. J Inst Eng (India): Ser B 94(4):247–256
Mahdavi S, Rahnamayan S, Deb K (2018) Opposition based learning: a literature review. Swarm Evolut Comput 39:1–23
Zhang Y (2012) Support vector machine classification algorithm and its application. In: International conference on information computing and applications. Springer, Berlin, pp 179–186
Haykin SS (2016) Neural networks and learning machines, 3rd edn. Pearson Education, London
Cover T, Hart P (1967) Nearest neighbor pattern classification. IEEE Trans Inf Theory 13(1):21–27
Rosenblatt F (1958) The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev 65(6):386
Altman NS (1992) An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat 46(3):175–185
Tizhoosh HR (2005) Opposition-based learning: a new scheme for machine intelligence. In: International conference on computational intelligence for modelling, control and automation and international conference on intelligent agents, web technologies and internet commerce (CIMCA-IAWTIC’06), vol 1. IEEE, USA, pp 695–701. https://doi.org/10.1109/CIMCA.2005.1631345
Hussain E, Mahanta LB, Borah H, Das CR (2020) Liquid based-cytology pap smear dataset for automated multi-class diagnosis of pre-cancerous and cervical cancer lesions. Data Brief 30:105589. https://doi.org/10.1016/j.dib.2020.105589
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE, USA, pp 770–778
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE, USA, pp 1–9
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint. arXiv:1409.1556
Buddhavarapu VG et al (2020) An experimental study on classification of thyroid histopathology images using transfer learning. Pattern Recognit Lett 140:1–9
Arun Kumar R, Vijay Franklin J, Koppula N (2022) A comprehensive survey on metaheuristic algorithm for feature selection techniques. Mater Today: Proc 64:435–441. International Conference on Advanced Materials for Innovation and Sustainability
Agrawal P, Abutarboush HF, Ganesh T, Mohamed AW (2021) Metaheuristic algorithms on feature selection: a survey of one decade of research (2009–2019). IEEE Access 9:26766–26791
Cinar AC, Kiran MS (2018) Similarity and logic gate-based tree-seed algorithms for binary optimization. Comput Ind Eng 115:631–646
Kiran MS, Gündüz M (2013) XOR-based artificial bee colony algorithm for binary optimization. Turk J Electr Eng Comput Sci 21:2307–2328
Malik MM, Haouassi H (2022) Efficient sequential covering strategy for classification rules mining using a discrete equilibrium optimization algorithm. J King Saud Univ Comput Inf Sci 34(9):7559–7569
Haouassi H, Mahdaoui R, Chouhal O, Bekhouche A (2022) An efficient classification rule generation for coronary artery disease diagnosis using a novel discrete equilibrium optimizer algorithm. J Intell Fuzzy Syst 43(3):2315–2331
Kennedy J, Eberhart R (1995) Particle swarm optimization. In: Proceedings of ICNN’95-international conference on neural networks, vol 4. IEEE, USA, pp 1942–1948. https://doi.org/10.1109/ICNN.1995.488968
Erlich I, Venayagamoorthy GK, Worawat N (2010) A mean-variance optimization algorithm. In: IEEE congress on evolutionary computation. IEEE, USA, pp 1–6. https://doi.org/10.1109/CEC.2010.5586027
Mirjalili S, Mirjalili SM, Lewis A (2014) Grey wolf optimizer. Adv Eng Softw 69:46–61
Mirjalili S (2015) Moth-flame optimization algorithm: a novel nature-inspired heuristic paradigm. Knowl Based Syst 89:228–249
Mirjalili S, Lewis A (2016) The whale optimization algorithm. Adv Eng Softw 95:51–67
Gandomi AH, Yang X-S, Alavi AH (2011) Mixed variable structural optimization using firefly algorithm. Comput Struct 89(23–24):2325–2336
Yang X-S, Gandomi AH (2012) Bat algorithm: a novel approach for global engineering optimization. Eng Comput. 29(5): 464–483. https://doi.org/10.1108/02644401211235834
De Jong KA (1975) Analysis of the behavior of a class of genetic adaptive systems. PhD thesis, University of Michigan Department, 72 Ann Arbor, MI, United States
Holland JH et al (1992) Adaptation in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence. MIT Press, Cambridge
Kiran MS (2015) The continuous artificial bee colony algorithm for binary optimization. Appl Soft Comput 33:15–23
Siegel S, Castellan N (1988) The Friedman two-way analysis of variance by ranks. Nonparametric statistics for the behavioral sciences, pp 174–184
Acknowledgements
The work is supported by SERB (DST), Govt. of India (Ref# EEQ/2018/000963). Authors are also thankful to Akil Kadolia and Soumyajyoti Dey for their efforts during the development of different modules.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflicts of interest.
Ethical approval
The article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Das, N., Mandal, B., Santosh, K. et al. Cervical cancerous cell classification: opposition-based harmony search for deep feature selection. Int. J. Mach. Learn. & Cyber. 14, 3911–3922 (2023). https://doi.org/10.1007/s13042-023-01872-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-023-01872-z