Abstract
Purpose
Cancer of unknown primary (CUP) accounts for 2–5% of all cancer diagnoses, wherein standard investigations fail to reveal the original tumor site. Basket trials allocate targeted therapeutics based on actionable somatic mutations, independent of tumor entity. These trials, however, mostly rely on variants identified in tissue biopsies. Since liquid biopsies (LB) represent the overall tumor genomic landscape, they may provide an ideal diagnostic source in CUP patients. To identify the most informative liquid biopsy compartment, we compared the utility of genomic variant analysis for therapy stratification in two LB compartments (circulating cell-free (cf) and extracellular vesicle (ev) DNA).
Methods
CfDNA and evDNA from 23 CUP patients were analyzed using a targeted gene panel covering 151 genes. Identified genetic variants were interpreted regarding diagnostic and therapeutic relevance using the MetaKB knowledgebase.
Results
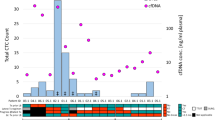
LB revealed a total of 22 somatic mutations in evDNA and/or cfDNA in 11/23 patients. Out of the 22 identified somatic variants, 14 are classified as Tier I druggable somatic variants. Comparison of variants identified in evDNA and cfDNA revealed an overlap of 58% of somatic variants in both LB compartments, whereas over 40% of variants were only found in one or the other compartment.
Conclusion
We observed substantial overlap between somatic variants identified in evDNA and cfDNA of CUP patients. Nonetheless, interrogation of both LB compartments can potentially increase the rate of druggable alterations, stressing the significance of liquid biopsies for possible primary-independent basket and umbrella trial inclusion.



Similar content being viewed by others
Data Availability
The datasets generated and analyzed during the current study are available from the corresponding authors on reasonable request.
Abbreviations
- AMP:
-
Association for Molecular Pathology
- ASCO:
-
American Society of Clinical Oncology
- CAP:
-
College of American Pathologists
- cfDNA:
-
Circulating cell-free DNA
- CNV:
-
Copy number variation
- CUP:
-
Cancer of unknown primary
- evDNA:
-
Extracellular vesicle DNA
- NGS:
-
Next-generation sequencing
- PBMC:
-
Peripheral blood mononuclear cells
- SNV:
-
Single-nucleotide variant
References
Binder C, Matthes KL, Korol D, Rohrmann S, Moch H. Cancer of unknown primary-Epidemiological trends and relevance of comprehensive genomic profiling. Cancer Med. 2018;7:4814–24.
Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379:1428–35.
Varadhachary GR, Raber MN. Carcinoma of unknown primary site. N Engl J Med. 2014;371:2040.
Blaszyk H, Hartmann A, Bjornsson J. Cancer of unknown primary: clinicopathologic correlations. APMIS. 2003;111:1089–94.
Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer. 2007;43:2026–36.
Hemminki K, Riihimaki M, Sundquist K, Hemminki A. Site-specific survival rates for cancer of unknown primary according to location of metastases. Int J Cancer. 2013;133:182–9.
Raghav K, et al. Cancer of unknown primary in adolescents and young adults: clinicopathological features, prognostic factors and survival outcomes. PLoS ONE. 2016;11: e0154985.
Fizazi K, et al. Cancers of unknown primary site: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v133–8.
Greco FA, Lennington WJ, Spigel DR, Hainsworth JD. Poorly differentiated neoplasms of unknown primary site: diagnostic usefulness of a molecular cancer classifier assay. Mol Diagn Ther. 2015;19:91–7.
Hainsworth JD, Fizazi K. Treatment for patients with unknown primary cancer and favorable prognostic factors. Semin Oncol. 2009;36:44–51.
Culine S. Prognostic factors in unknown primary cancer. Semin Oncol. 2009;36:60–4.
Losa F, et al. SEOM clinical guideline on unknown primary cancer (2017). Clin Transl Oncol. 2018;20:89.
Greco FA, Lennington WJ, Spigel DR, Hainsworth JD. Molecular profiling diagnosis in unknown primary cancer: accuracy and ability to complement standard pathology. J Natl Cancer Inst. 2013;105:782–90.
Varadhachary GR, et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J Clin Oncol. 2008;26:4442–8.
Murtaza M, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6.
Pennec SL, et al. Intratumor heterogeneity and clonal evolution in an aggressive papillary thyroid cancer and matched metastases. Endocr Relat Cancer. 2015;22:205–16.
Chaudhuri AA, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–403.
Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209.
Tie J, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92.
Forshew T, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4.
Fackler MJ, et al. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer Res. 2014;74:2160–70.
San Lucas FA, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2016;27:635–641.
Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86.
Kahlert C. Liquid biopsy: is there an advantage to analyzing circulating exosomal DNA compared to cfDNA or are they the same? Cancer Res. 2019;79:2462–5.
Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930.
Bernard V, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterol. 2019;156:108–118 e4.
Wagner AH, et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat Genet. 2020;52:448–57.
Castillo J, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol Off J Eur Soc Med Oncol. 2018;29:223–9.
Semaan A, et al. Defining the comprehensive genomic landscapes of pancreatic ductal adenocarcinoma using real-world endoscopic aspiration samples. Clin Cancer Res. 2021;27:1082–93.
Li MM, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagnostics. 2017;19:4–23.
Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562.
Jiao Y, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–3.
Zou S, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;(51)5:1–11.
Sallman DA, et al. Eprenetapopt (APR-246) and azacitidine in TP53-mutant myelodysplastic syndromes. J Clin Oncol. 2021;39:1584–94.
Lindemann A, et al. COTI-2, a novel thiosemicarbazone derivative, exhibits antitumor activity in HNSCC through p53-dependent and -independent mechanisms. Clin Cancer Res. 2019;25:5650–62.
Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. 2011;364:2507–2516. https://doi.org/10.1056/NEJMoa1103782.
Solomon BJ, et al. First-line crizotinib versus chemotherapy in ALK -positive lung cancer. N Engl J Med. 2014;371:2167–77.
Hainsworth JD, et al. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J Clin Oncol. 2013;31:217–23.
Yoon HH, et al. Gene expression profiling identifies responsive patients with cancer of unknown primary treated with carboplatin, paclitaxel, and everolimus: NCCTG N0871 (alliance). Ann Oncol. 2016;27:339–44.
Hayashi H, et al. Randomized phase II trial comparing site-specific treatment based on gene expression profiling with carboplatin and paclitaxel for patients with cancer of unknown primary site. J Clin Oncol. 2019;37:570–9.
Fizazi K, et al. A phase III trial of empiric chemotherapy with cisplatin and gemcitabine or systemic treatment tailored by molecular gene expression analysis in patients with carcinomas of an unknown primary (CUP) site (GEFCAPI 04). 2019. https://doi.org/10.1093/annonc/mdz394.
Woodcock J, LaVange LM. Master protocols to study multiple therapies, multiple diseases, or both. N Engl J Med. 2017;377:62–70.
Pauli C, et al. A challenging task: identifying patients with cancer of unknown primary (CUP) according to ESMO guidelines: the CUPISCO trial experience. Oncologist. 2021;26: e769.
Wolfers J, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303.
Kato S, et al. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res. 2017;77:4238–46.
Nguyen L, Van Hoeck A, Cuppen E. Machine learning-based tissue of origin classification for cancer of unknown primary diagnostics using genome-wide mutation features. Nat Commun. 2022;13:1–12.
Laprovitera N, et al. Genetic characterization of cancer of unknown primary using liquid biopsy approaches. Front Cell Dev Biol. 2021;9.
Acknowledgements
We thank our patients and their families. We dedicate this study to the memory of our beloved colleague Dr. Gauri R. Varadhachary who was responsible for initiating this study and was always there for her patients.
Funding
A.M. is supported by the MD Anderson Pancreatic Cancer Moon Shot Program, the Sheikh Khalifa bin Zayed Al-Nahyan Foundation, and the NIH (R01CA218230, U01CA196403, U01CA200468, and P50CA221707). V. Bernard is supported by the NIH (U54CA096300, U54CA096297, and T32CA217789). J.J.L. is supported by the NIH (T32CA009599). A.S. is supported by the German Research Foundation (SE-2616/2–1).
Author information
Authors and Affiliations
Contributions
F.R.K. performed experiments and data analysis and wrote the manuscript, B.M.S. performed experiments, V. Branchi conducted data analysis, V. Bernard and J.J.L. edited the manuscript. K.P.S.R. helped with clinical annotations, A.M. provided funding and scientific guidance and edited the manuscript. A.S. and P.A.G, conceptualized and guided the work and edited the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
A.M. receives royalties for a pancreatic cancer biomarker test from Cosmos Wisdom Biotechnology, and this financial relationship is managed and monitored by the UTMDACC Conflict of Interest Committee. A.M. is also listed as an inventor on a patent that has been licensed by Johns Hopkins University to Thrive Earlier Detection (an Exact Sciences Company). A.M. is a consultant for Freenome and Tezcat. All other authors declare no conflicts of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance
Analysis of genomic variants in liquid biopsies has potential as a diagnostic tool for CUP samples. We compared cfDNA and evDNA by a targeted NGS approach and identified druggable targets that could help to stratify patients for inclusion into clinical trials.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kolbinger, F.R., Bernard, V., Lee, J.J. et al. Significance of Distinct Liquid Biopsy Compartments in Evaluating Somatic Mutations for Targeted Therapy Selection in Cancer of Unknown Primary. J Gastrointest Canc 54, 1276–1285 (2023). https://doi.org/10.1007/s12029-023-00922-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-023-00922-7