Abstract
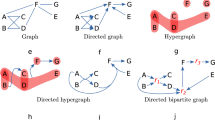
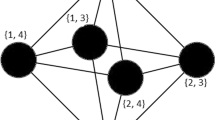
The analysis of the structure of chemical reaction networks is crucial for a better understanding of chemical processes. Such networks are well described as hypergraphs. However, due to the available methods, analyses regarding network properties are typically made on standard graphs derived from the full hypergraph description, e.g. on the so-called species and reaction graphs. However, a reconstruction of the underlying hypergraph from these graphs is not necessarily unique. In this paper, we address the problem of reconstructing a hypergraph from its species and reaction graph and show NP-completeness of the problem in its Boolean formulation. Furthermore we study the problem empirically on random and real world instances in order to investigate its computational limits in practice.
Similar content being viewed by others
References
Limboole. http://fmv.jku.at/limboole/index.html. Institute for Formal Models and Verification (2013)
Alon, N., Galil, Z., Margalit, O., Naor, M.: Witnesses for boolean matrix multiplication and for shortest paths. In: Foundations of Computer Science, 1992. Proceedings of 33rd Annual Symposium, pp. 417–426. IEEE (1992)
Andersen, J., Flamm, C., Merkle, D., Stadler P.: Maximizing output and recognizing autocatalysis in chemical reaction networks is NP-complete. J. Syst. Chem. 3(1) (2012)
Barrett, C., Stump, A., Tinelli, C.: The SMT-LIB Standard: Version 2.0. In: Gupta, A., Kroening, D. (eds.) Proceedings of the 8th International Workshop on Satisfiability Modulo Theories (Edinburgh, UK) (2010)
Benkö G., Centler F., Dittrich P., Flamm C., Stadler B.M.R., Stadler P.F.: A topological approach to chemical organizations. Alife 15, 71–88 (2009)
Biere, A.: Lingeling, plingeling, picosat and precosat at sat race 2010. FMV Report Series Technical Report 10(1) (2010)
Biere, A., Heule, M., van Maaren, H., Walsh, T.: Handbook of Satisfiability, Frontiers in Artificial Intelligence and Applications, vol. 185 (2009)
Borenstein, E., Kupiec, M., Feldman, M.W., Ruppin, E.: Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc. Natl. Acad. Sci. USA 105, 14482–14487 (2008)
Capobianco M., Molluzzo J.C.: Examples and counterexamples in graph theory. J. Graph Theory 2(3), 274–274 (1978)
Cottret, L., Wildridge, D., Vinson, F., Barrett, M.P., Charles, H., Sagot, M.-F., Jourdan, F.: Metexplore: a web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res. 38(suppl 2), W132–W137 (2010)
Craciun G., Feinberg M.: Multiple equilibria in complex chemical reaction networks: II. The species-reaction graph. SIAM J. Appl. Math. 66, 1321–1338 (2006)
De Moura L., Bjørner N.: Z3: An efficient SMT solver. Tools Algorithms Constr. Anal. Syst. (TACAS’) 08(4963), 337–340 (2008)
Domijan M., Kirkilionis M.: Graph theory and qualitative analysis of reaction networks. Netw. Heterog. Media 3, 295–322 (2008)
Eén, N., Sörensson, N.: An extensible sat-solver. In: Theory and Applications of Satisfiability Testing, pp. 333–336. Springer, New York (2004)
Erdős P., Rényi A.: On the evolution of random graphs. Magyar Tud. Akad. Mat. Kutató Int. Közl 5, 17–61 (1960)
Feinberg M.: Complex balancing in general kinetic systems. Arch. Ration. Mech. Anal. 49, 187–194 (1972)
Gallo G., Longo G., Pallottino S., Nguyen S.: Directed hypergraphs and applications. Discrete Appl. Math. 42(2), 177–201 (1993)
Garey, M., Johnson, D.: Computers and Intractability: A Guide to the Theory of NP-Completeness (1979)
Han J.D.J., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J., Cusick M.E., Roth F.P. et al.: Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430(6995), 88–93 (2004)
Horn F.J.M., Jackson R.: General mass action kinetics. Arch. Ration. Mech. Anal. 47, 81–116 (1972)
Ivanova A.N.: Conditions for uniqueness of stationary state of kinetic systems, related, to structural scheme of reactions. Kinet. Katal 20, 1019–1023 (1979)
Jeong H., Tombor B., Albert R., Oltvai Z., Barabási A.: The large-scale organization of metabolic networks. Nature 407(6804), 651–654 (2000)
Kaltenbach, H.-M.: A unified view on bipartite species-reaction and interaction graphs for chemical reaction networks. Technical Report 1210.0320, arXiv (2012)
Klamt S, Haus U-U, Theis F (2009) Hypergraphs and cellular networks. PLoS Comput Biol 5:e1000385
Lacroix V., Cottret L., Thébault P., Sagot M.: An introduction to metabolic networks and their structural analysis. IEEE/ACM Trans. Comput. Biol. Bioinforma. 5(4), 594–617 (2008)
McNaught, A., Wilkinson, A.: IUPAC. Compendium of Chemical Terminology, vol. 2. Blackwell Scientific Publications, Oxford (1997)
Miettinen P., Mielikainen T., Gionis A., Das G., Mannila H.: The discrete basis problem. IEEE Trans. Knowl. Data Eng. 20(10), 1348–1362 (2008)
Mincheva M., Roussel M.R.: A graph-theoretic method for detecting potential Turing bifurcations. J. Chem. Phys 125, 204102 (2006)
Mincheva M., Roussel M.R.: Graph-theoretic methods for the analysis of chemical and biochemical networks. I. Multistability and oscillations in ordinary differential equation models. J. Math. Biol. 55, 61–68 (2007)
Shinar G., Feinberg M.: Concordant chemical reaction networks and the species-reaction graph. Math. Biosci. 241, 1–23 (2013)
Skiena, S.: Implementing Discrete Mathematics: Combinatorics and Graph Theory with Mathematica (1990)
Sörensson, N., Eén, N.: Minisat v1. 13-a sat solver with conflict-clause minimization. SAT 2005, 53 (2005)
Soulé C.: Graphic requirements for multistationarity. ComplexUs 1, 123–133 (2003)
Tanaka R.: Scale-rich metabolic networks. Phys. Rev. Lett. 94(16), 168101 (2005)
Thomas, R.: On the relation between the logical structure of systems and their ability to generate multiple steady states or sustained oscillations. In: Della Dora, J., Demongeot, J., Lacolle, B. (eds.) Numerical Methods in the Study of Critical Phenomena, vol. 9, pp. 180–193. Springer, Berlin (1981)
Tseitin, G.: On the complexity of derivation in propositional calculus. In: Slisenko, A.O. (ed.) Structures in Constructive Mathematics and Mathematical Logic, Part II, Seminars in Mathematics (translated from Russian), Steklov Mathematical Institute, pp. 115–125 (1968)
Tseitin G.: On the complexity of derivation in propositional calculus. Autom. Reason. Class. Papers Comput. Logic 2, 466–483 (1983)
Volpert, A., Ivanova, A.N.: Mathematical models in chemical kinetics. In: Mathematical Modeling, pp. 57–102. Nauka, Moscow (1987) (Russian)
Wagner A., Fell D.A.: The small world inside large metabolic networks. Proc. R. Soc. Lond. B 268, 1803–1810 (2001)
Watts D.J., Strogatz S.H.: Collective dynamics of ‘small-world’ networks. Nature 393, 409–410 (1998)
Zeigarnik, A.V.: On hypercycles and hypercircuits in hypergraphs. In: Hansen, P., Fowler, P.W., Zheng, M. (eds.) Discrete Mathematical Chemistry, volume 51 of DIMACS Series in Discrete Mathematics and Theoretical Computer Science, pp. 377–383. American Mathematical Society, Providence (2000)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fagerberg, R., Flamm, C., Merkle, D. et al. On the Complexity of Reconstructing Chemical Reaction Networks. Math.Comput.Sci. 7, 275–292 (2013). https://doi.org/10.1007/s11786-013-0160-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11786-013-0160-y