Abstract
Acute myocardial infarction (AMI) or heart attack is a significant global health threat and one of the leading causes of death. The evolution of machine learning has greatly revamped the risk stratification and death prediction of AMI. In this study, an integrated feature selection and machine learning approach was used to identify potential biomarkers for early detection and treatment of AMI. First, feature selection was conducted and evaluated before all classification tasks with machine learning. Full classification models (using all 62 features) and reduced classification models (using various feature selection methods ranging from 5 to 30 features) were built and evaluated using six machine learning classification algorithms. The results showed that the reduced models performed generally better (mean AUPRC via random forest (RF) algorithm for recursive feature elimination (RFE) method ranges from 0.8048 to 0.8260, while for random forest importance (RFI) method, it ranges from 0.8301 to 0.8505) than the full models (mean AUPRC via RF: 0.8044). The most notable finding of this study was the identification of a five-feature model that included cardiac troponin I, HDL cholesterol, HbA1c, anion gap, and albumin, which had achieved comparable results (mean AUPRC via RF: 0.8462) as to the models that containing more features. These five features were proven by the previous studies as significant risk factors for AMI or cardiovascular disease and could be used as potential biomarkers to predict the prognosis of AMI patients. From the medical point of view, fewer features for diagnosis or prognosis could reduce the cost and time of a patient as lesser clinical and pathological tests are needed.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
AMI or heart attack is one of the acute coronary syndromes (ACS) which is a consequence of the sudden loss of blood supply to the heart muscle due to partial or complete blockage of a coronary artery. Through the electrocardiogram (ECG) findings, AMI is usually classified into ST-segment elevated myocardial infarction (STEMI) and non-ST-segment elevated myocardial infarction (NSTEMI). A STEMI occurs when there is a complete blockage in the coronary artery and would show significant changes in the ST-segment on the ECG. In contrast, NSTEMI has a partial blockage in the coronary artery and would not show any change in the ST-segment on the ECG. STEMI patients are having a greater risk of death than NSTEMI patients [1].
Various risk factors may cause AMI including smoking, hypertension, high body mass index (BMI), hyperglycaemia, dyslipidaemia (due to an unhealthy diet), alcohol and/or drugs harmful use, and physical inactivity [2]. Studies had shown that non-communicable diseases and psychological, genetic, and environmental factors also can affect AMI patients especially post-myocardial infarction (post-MI). Demographic parameters such as gender, age, family history of having cardiovascular diseases (CVD), ethnicity, and socio-economy other than comorbidities and air pollution or beliefs likewise influence AMI mortality too [3, 4].
According to World Health Organization (2021) and International Health Metric Evaluation (2020), an estimation of 18.6 million people died from CVD in the year 2019 and 85% of these mortalities were due to AMI and stroke. Over 75% of these deaths occurred in low- and middle-income countries [2, 5]. In Malaysia, ischemic heart disease (IHD), another term for AMI, is listed as the principal cause of death which made up 15% of all death in the year 2019 with nearly 70% of IHD death being male [6]. The percentage slightly dropped in 2021 to 13.7% due to peaking in death from Covid-19 infection which made up 19.8% [7]. Despite that, AMI studies in low- and middle-income countries are still small against those in developed countries [8]. Hence, it is important to carry out studies related to this chronic disease such as risk classification and discovering potential biomarkers for early diagnosis and prognosis.
1.1 Background study
Currently, traditional risk classification models are still the gold standard and are widely utilized in CVD studies. Some commonly used conventional models are Thrombolysis in Myocardial Infarction (TIMI) [9, 10], Framingham Risk Score (FRS) [11, 12], Global Registry of Acute Coronary Events (GRACE) [13, 14], and History, ECG, Age, Risk factors and Troponin (HEART) [15]. The selection of these conventional models is influenced by the features used in the models. Some architectures required straightforward features (e.g., FRS), while some may require complex pathological results (e.g., GRACE and HEART). In addition, all the conventional risk classification models were developed based on the Caucasian cohort; thus, some adaptations might be needed to modify these models to be more suited for other ethnicities. Nevertheless, these conventional models provided a simple and quick approach where limited resources are available.
Precision and personalized medicine, along with improving risk stratification, especially in CVD medicine, have led to the study and proposal of multiple cardiac and non-cardiac biomarkers. Creatine kinase - myocardial banding (CKMB) and cardiac troponins (cTn: cTnI or cTnT) are among the most commonly used biomarkers to diagnose and stratify AMI patients, according to the review by Aydin et al. (2019) [16] and the Universal Definition of AMI [17].
In addition to CKMB and cTn, other biomarkers have been proposed, including molecular description, mechanism of action, and activity level relative to the disease, involving lipids, salivary, and urine biomarkers, apart from common blood test components [16, 18].
Machine learning (ML) is a division of artificial intelligence (AI) that uses data and algorithms to mimic the way that humans learn and solve problems and improve their accuracy through learning [19]. The emergence of ML has greatly contributed to the field of AMI risk classification. Some commonly used ML algorithms in AMI or CVD studies are logistic regression (LR), support vector machine (SVM), k-nearest neighbour (KNN), artificial neural network (ANN), and random forest (RF) [20,21,22,23,24,25]. ML can be divided into three types of learning namely supervised learning, unsupervised learning, and reinforcement learning. In supervised learning, the algorithms are trained using labelled data. Conversely, for unsupervised learning models, algorithms are trained against unlabeled data. Meanwhile, reinforcement learning is trained based on the rewarding behaviours in an environment and provided feedback to improve the learning process.
Literature studies had proved that ML models outperformed conventional risk classification models. For example, Alaa et al. [26] proved that the proposed ML model outperformed FRS and Cox PH models. Their ML model included new predictors such as individuals with a history of diabetes, which were not usually used in conventional models and showed improvement in risk classification of relevant subpopulations. Another 1-year mortality classification study by Sherazi et al. [27] had shown that ML models outperformed GRACE in patients with the ACS. Moreover, ML models also showed better performance than the TIMI model in short- and long-term mortality predictions for STEMI as TIMI scores had underestimated patients’ risks of mortality in the study [28]. Hence, ML could serve as a better choice than conventional methods in risk classification for CVD studies, as ML could identify hidden patterns and include various types of data, whereas conventional methods are mainly used to identify the causality between limited variables [8].
1.2 Contribution of this work
The objective of this study is to develop an integrated machine learning and biomarker-based prognostic model for AMI patients. The classification ability of full models (all features) was compared with the reduced models (selected features) using supervised ML algorithms. Feature selections were implemented to select the optimum features subset for the reduced models before being fed into the ML classifiers. Next, common clinicopathologic features selected among the best two feature selection methods were identified and the findings were validated with the literature studies. These optimum features identified could be used as potential biomarkers for the early detection or treatment of AMI patients.
Our contributions are as follows:
-
1)
We proposed an integrated model for AMI using a feature selection and machine learning approach for biomarker discovery. This approach reduces the time required for interpretation and interpolation of heterogeneous medical data compared to manual and conventional approaches, which aids faster classification for decision-making and risk stratification.
-
2)
We compared various methods of feature selection to identify the optimal subset of features for in-hospital mortality classification including filter, wrapper, and embedded methods. Other AMI/CVD studies typically focused on comparing results from different classification algorithms (ANN vs SVM vs KNN, etc.) without comparing the results from different feature selection methods.
-
3)
After conducting the feature selection task using wrapper and embedded, we identified five common features between the best two selection models. These features have the potential to serve as biomarkers for early diagnosis and prognosis of AMI. We verified these features with literature reviews, which confirmed that they are associated with AMI and consider as important markers.
-
4)
To address the issue of imbalanced data classification, we chose the area under the Precision-Recall curve (AUPRC) as our primary performance measure, instead of the commonly used area under the Receiver Operating Characteristic curve (AUROC). Medical data often have skewed and imbalanced datasets that can affect the performance of the classification models. Therefore, we used AUPRC and stratified shuffle - split to improve the accuracy of our results and mitigate or avoid class imbalance issues.
2 Methods
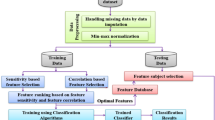
This research was approved by the Medical Research Ethics Committee, University of Malaya Medical Centre (UMMC) with MREC ID NO.: 201985–7712. The proposed framework for the AMI biomarker discovery using an integrative approach of machine learning and feature selection is shown in Fig. 1.
The proposed framework for biomarker discovery in AMI using machine learning and feature selection. Abbreviations: AMI, acute myocardial infarction; ANN, artificial neural network; AUPRC, area under the precision-recall curve; DT, decision tree; kNN, k-nearest neighbours; LR, logistic regression; PCC, Pearson’s coefficient correlation; RF, random forest; RFE, recursive feature elimination; RFI, random forest importance; SVM, support vector machine (refer to Appendix A for the full list of abbreviations)
2.1 Data collection
The AMI dataset was collected from the Department of Pathology, UMMC. The dataset collected consisted of five demographical data (features) and 84 clinicopathologic data (features) for 140 AMI patients who were admitted to the hospital UMMC between December 2019 and June 2020. The data were collected from various clinical and pathological reports including complete blood count (CBC), lipid profile (LP), differential count (DC), renal function test (RFT), liver function fest (LFT), diabetes, coagulation test (CT), and cardiac biomarkers test (CB). Table 1 shows a summary of the demographic characteristics of 140 AMI patients.
2.2 Hardware and software
The computer hardware used to perform all the processing and computational work is a laptop with Intel core i5 1.6 Ghz CPU processor and 8 GB RAM. The operating system is 64-bit Windows 10/11.
The programs were built and run using Python 3.8.10 and IPython 7.32.0 through SPyDEr version 4.1.4. The Python libraries used are NumPy, Pandas, matplotlib, Scikit-Learn (sklearn), Keras, and TensorFlow.
2.3 Data preprocessing
Initially, there were 89 features in the dataset. The percentage of the missing values for each of the features was calculated, and those features with missing values of more than 50% were removed from the dataset, a standard data preprocessing technique as suggested by Rengaraju [29]. Hence, only 62 features remained after the data-cleansing step. All 62 features and the proportion of missing values are listed in Appendix B. However, there were still 670 missing values in the remaining 8680 data points (7.72%) among the 62 features. Hence, data imputation is needed to fill in these missing values. A summary of the data description is listed in Table 2.
Data imputation was computed using median imputation as all the missing features are continuous data. The median was chosen as it has a better representation when the feature has a skewed distribution [30]. The median value for each feature would be computed and missing values within the same feature would be imputed with the same median value. After that, data normalization was done by using MinMaxScaler which rescaled all the continuous features to a range of 0 to 1. This method preserves the shape of the original distribution without changing the information embedded in the original data. Besides, when the features are relatively smaller or closer to the normal distribution, the algorithm could converge faster [31]. The normalization step can be represented by Eq. (1) below:
where Xi is the value of observation in the feature, and Xmin and Xmax are the minimum value and maximum value for the feature, respectively. No normalization was needed on the categorical features.
2.4 Feature selection
Three types of feature selection were used in this study which are (1) filter method — Pearson’s coefficient correlation (PCC); (2) wrapper method — recursive feature elimination (RFE); and (3) embedded method — random forest importance (RFI).
PCC’s values are ranged from + 1 to − 1, where + 1 indicates a total positive correlation and − 1 indicates a total negative correlation, and 0 represents no correlation between the variables [32, 33]. PCC can be calculated by using Eq. (2) below:
where cov(x,y) is the covariance of the input feature and the target feature (in-hospital mortality), and \({\sigma }_{x}\) and \({\sigma }_{y}\) represent the standard deviation of the input feature and the target feature, respectively.
Whereas RFE method evaluates the variables in subsets and uses the heuristic search methods to obtain an optimal subset, RFE performs a greedy search to find the best-performing variable subset by removing the features until the optimum number of features is identified and ranks the features based on the order of their elimination [34].
RFI uses the importance score of each feature based on the Gini to select the internal split points of a decision tree when training in RF. The higher the importance score indicates the more optimal the feature. In this study, the important score was computed as the mean and standard deviation of accumulation of the impurity decrease within each tree. The importance score is measured by observing how the impurity of the split for each feature is decreasing, and the feature with the highest decrement of impurity will be selected until the optimum subset of features is chosen [35].
2.5 Classification
Six supervised ML methods were used as the classifiers in this study, namely decision tree (DT), random forest (RF), k-nearest neighbours (KNN), artificial neural network (ANN), support vector machine (SVM), and logistic regression (LR). The classifiers from the Python Scikit-learn (sklearn) library and Keras library were used.
DT starts from the root node, and the tree splits into branches (decision nodes). This process continues until the end of the branches which are the leaf nodes that cannot be split further [36]. In this study, Gini impurity was set to measure the quality of a split in DT and the minimum number of samples at the leaf node (min_samples_leaf) was set to 10.
RF is an ensemble learning algorithm that consists of many DT to provide solutions to complex problems and also improve the model performance. In this study, the values for number of trees (n_estimators) from 50 to 200 were tested and 100 was selected as the optimal value. Gini impurity was used to measure the quality of a split due to its simplicity [37].
KNN works by calculating the distance between the unknown samples and the data points [38]. The class of the unknown sample is determined by ‘majority voting’ from the labels of k-nearest data points. A general rule of thumb in choosing the k value is k = square root (N), where N = number of samples. In this study, k = square root(140) = 11.8 and only the odd numbers were selected for k to avoid any ties in classification. Hence, k values that range from k = 3, 5, 7, 9, 11 were tested and k = 5 yielded the optimum results. The uniform weight was used where all points in each neighbourhood were weighted equally.
ANN is a biological-inspired computational network that learns through the interconnected neurons in the layered architecture which resembles the human brain [39]. In this study, we adopted the multi-layer perceptron (MLP) due to its simplicity and small dataset. The ANN architecture adopted in this study consisted of two hidden layers with 80 and 40 hidden neurons respectively with rectified linear unit (ReLU) as the activation function. The activation function for the output layer was set to sigmoid for a binary classification. Besides that, other parameters such as epochs = 30 and batch size = 10 were set for this study.
The SVM is used to find the optimal hyperplane that could classify the classes well in an N-dimensional space, where N is the number of features [40]. In this study, some commonly used kernel functions such as linear, polynomial, and radial basis functions (RBF) were tested, and the RBF kernel was set as the optimal kernel in this study.
LR is used to model the relationship between one or more independent variables and a dependent variable with a linear equation [41]. The liblinear solver (library for large linear classification) was used in this study as it is suitable for small datasets. The optimal hyper-parameters and the Python libraries used in this study are summarized in Table 3.
In this study, firstly, all 62 features were used and trained in the full model classification as the input to predict the in-hospital mortality of AMI patients. Next, feature selection methods were applied to the 62 features to select the optimum features. Lastly, common features selected from the best two feature selection models were used as the reduced model in the ML classification step. For the model development, a 10-time repeated five-fold cross-validation (5-CV) stratified-shuffle split method was used. The stratified split method ensures each set of data contains a similar percentage of samples for each class and thus avoids class imbalance problems.
2.6 Performance evaluation
The confusion matrix was adopted to define the performance of the classification model. The confusion matrix is a N × N matrix that compares the actual values with the predicted values. It summarizes the number of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). The model performance such as testing accuracy, precision, recall, and F1 score (Eqs. 3–6) were measured using these four values obtained from the confusion matrix. Precision is the ratio between the TP and all the positive classes, while recall quantifies the amount of TP out of all positive examples in the dataset. F1 score gives a harmonic mean that balances precision and recall [42]. The equations for testing accuracy, precision, recall and F1 score were shown in Eqs. 3 to 6, respectively.
Next, the precision-recall curve (PRC) was plotted with the precision values as the y-axis and the recall values as the x-axis [43]. The area under the PRC (AUPRC) was calculated. In an imbalanced classification problem with two classes, the positive class is always referred to as the minority class. In this study, the “died” class is positive). According to Fu et al. [44], the PRC is more suitable than the receiver operating characteristic (ROC) curve as the performance measurement for the imbalanced datasets due to both the precision and the recall being focused on the positive class only. Thus, this makes the PRC an effective assessment tool for imbalanced classification models.
The precision and recall values were computed from the testing set and the average AUPRC of each model was calculated. Value ranges of AUPRC are between 0 and 1. The higher the AUPRC, the better the model performs in classifying in-hospital mortality of patients from the clinicopathologic data. The baseline of AUPRC is equal to the fraction of positive class (0.429 in this study), calculated using Eq. 7 below:
Hence, an AUPRC that is lower or near 0.429 is considered a no-skill classifier that cannot discriminate between the classes [45]. In this study, the model that acquired the best AUPRC based on the testing dataset was selected as the best model.
3 Results
In the full model classification, all 62 features were used to train and test with the six classifiers. Table 4 shows the results of full models with 5-CV. Each model was run 10 times, and the average testing accuracy, AUPRC, and F1 score were taken.
In Table 4, the classifier that showed the best performance in full models was RF (accuracy = 74.93%, AUPRC = 0.8044). Conversely, the worst performance was obtained by KNN with an accuracy of 59.07% and an AUPRC of 0.5351. However, in terms of training time, ANN took the longest time with 21.4998 s while LR took the shortest time with 0.11 s.
Next, three feature selection methods were implemented to build the reduced models. Feature selection using Pearson’s coefficient correlation (PCC) was first performed with the selection of 30 features, which is about 50% of the full model (62 features). Next, PCC was continued with the reduced number of features until the best model was obtained. Table 5 shows the classification results with PCC feature selection with 5-CV.
In Table 5, it can be observed that 15 features filtered using PCC achieved the highest AUPRC of 0.7450 with RF. Nonetheless, the overall model performance did not improve if compared to the full models.
On the other hand, feature selection using recursive feature elimination (RFE) and random forest importance (RFI) was initiated with the selection of a 15-feature subset removing the feature one by one until the best result was obtained. Next, common features selected among the best models from each method (RFE and RFI) were identified, and these common features were used in a more reduced model to classify the patients. Table 6 shows the results of RFE and RFI models from 15 to 11 features.
In Table 6, RF performed the best if compared to other classifiers with accuracy > 76% and AUPRC > 0.80. The best results were achieved by RFE-13 features (Accuracy = 76.72%, AUPRC = 0.8260) for the wrapper method and RFI-13 features (Accuracy = 78.57, AUPRC = 0.8505) for the embedded method. However, RFI-13 features model performed slightly better than the RFE-13 features model. Furthermore, it can be observed that the performance of most of the models achieved AUPRC > 0.7. Overall, the performance of reduced models with RFE and RFI is better than the performance of PCC and the full models for all the classifiers. Figure 2 shows one of the confusion matrices and PRC computed from one of the 5-CV runs in the RFI-13 features model with RF.
The time taken for three of the best feature selection models is computed and compared in Table 7. PCC with 15 features took the longest time to select features, while RFI-13 had the shortest feature selection time.
Next, common features selected by the two best feature selection models (RFE and RFI) were identified. Five common features were found and extracted into the more reduced model for the classification. Table 8 shows the list of common features selected, while Table 9 shows the classification results from the five common features.
The performance of the 5-feature model was better than the full model and was comparable to both the RFE-13 features model and RFI-13 features model with the best accuracy of 79.22% and the best AUPRC of 0.8462 on the RF classifier. Overall, RF outperformed the other classifiers in both full models and reduced models with AUPRC > 0.80 except in the reduced model with PCC. In terms of time consumption, the training time was the longest for ANN with 1.7286 s and the shortest via DT classifier with 0.0780 s.
4 Discussion
In this study, three performance measures of the classification models were taken and AUPRC was used as the top measure to select the best model. Accuracy was not chosen as the top measure since accuracy may be biased towards the majority/dominance class as the dataset is imbalanced in this study.
For full models and reduced models’ classification, RF outperformed other classifiers by achieving the best AUPRC. RF is a bagging algorithm in which bootstrapping enables RF to work well on relatively small datasets [46]. The performance of the reduced models is better (except in PCC) than the full models as the presence of some noisy features in the models (PCC models) caused overfitting and reduced the models’ performance. Overfitting may occur where some of the noisy features entered into the model simply by chance [47].
The implementation of PCC did not promote the performance of the reduced models. The performances dropped slightly (Table 5) if compared to the performance of the full models (Table 4). The disadvantage of filter methods is the ignorance of feature dependencies as each feature is considered an independent feature [47]. In this study, clinical and pathological features are related and have impacts on each other in the in-hospital mortality classification.
Nevertheless, the model performance of the reduced models increased after the implementation of RFE and RFI feature selection. This is due to some irrelevant features being eliminated, and the noise in the dataset had been reduced. As referred to in Table 6, it can be observed that RF achieved AUPRC > 0.80 for all the reduced models. RFE (wrapper) usually performed better than the PCC method (filter) as it can detect the interaction between the variables as well as identify the optimal feature subset [48, 49]. Similar to RFE, RFI (embedded) also considers the interaction of features. The tree-based strategies used in RFI rank by the improvements made to the internal node and identify the most important features by pruning trees below a particular node [50].
In Table 9, it can be observed that the performances of the 5-feature models were comparable with RFI-13 features and RFE-13 features models. The five common features selected are cardiac troponin I (cTnI), high-density lipoprotein cholesterol (HDL cholesterol), glycated haemoglobin (HbA1c), anion gap, and albumin. These five common features were further verified with the previous AMI studies, and all of them were proven to be important biomarkers in AMI.
4.1 Literature verification
Cardiac troponin I (cTnI) is a key regulatory protein in the cardiac that regulates the contractions of cardiac muscles. A troponin test measures the levels of cTnI proteins in the blood. These proteins will be released when the heart muscle has been damaged e.g. in a heart attack. A study that included 14,061 STEMI patients by Wanamaker et al. [51] proved that elevated admission troponin (both cTnI and cTnT) level is associated with higher mortality in STEMI patients. Likewise, Matetzky et al. [52] collected cTnI from 110 STEMI patients and discovered that patients with elevated cTnI were more likely to develop congestive heart failure (CHF) and death. cTnI are more sensitive than CKMB, while cTnT is poorer than CKMB for the diagnosis of AMI [53]. Due to the longer elevation period (1 to 2 weeks), cTnI is commonly used as a prognostic marker.
HDL cholesterol commonly known as “good cholesterol” absorbs excess cholesterol and takes it back to the liver where it is broken down and eliminated from the body. High levels of HDL cholesterol can reduce the risk of heart disease. In the study of Lee et al. [54] using samples of AMI patients enrolled in the Korea Acute Myocardial Infarction Registry (KAMIR), patients with decreasing HDL cholesterol showed significantly higher rates of 12-month major adverse cardiac events (MACE) as compared to the patients with increasing HDL cholesterol. Besides, Salonen et al. also confirmed that total HDL cholesterol and HDL2 (subfraction) levels have inverse associations with AMI risk, i.e., higher HDL may be a protective factor, while an increase in HDL3(subfraction) would increase AMI risk [55].
HbA1c develops when haemoglobin joins with glucose in the blood, becoming “glycated.” This is an important indicator especially for diabetes mellitus (DM) patients as higher HbA1c will increase the risk of developing diabetes-related complications. It provides a picture of the blood glucose level across a 3–6-month period. A study by Salinero-Fort et al. [56], with 114 cases of AMI, showed that patients with first AMI had higher values of HbA1c. Similarly, Pan et al. [57] in their systematic review proved that HbA1c is an important indicator for in-hospital mortality and short-term mortality classification in ACS patients without known DM and without DM.
The anion gap blood test is used to test whether blood has an imbalance of electrolytes, where acidosis indicates too much acid in the blood (high anion gap) and alkalosis indicates not enough acid in the blood (low anion gap). In a study by Sahu et al. [58], they revealed that in-hospital death was much higher in patients with initial anion gap acidosis (33%) if compared to patients with normal anion gap (8%). They also concluded that the admission anion gap is an important risk stratification indicator for AMI patients. Another study by Tang et al. [59] proved that 30-day and 90-day all-cause mortalities in patients with CHF (comorbidities included AMI) were associated with higher serum anion gap.
Albumin is a protein that is produced in the liver and helps to carry important substances enter the bloodstream. Albumin also helps to prevent fluids from leaking out of the bloodstream. Islam et al. [60] concluded that first-attack AMI patients with lower albumin (< 3.50 g/dl) had a worse in-hospital outcomes. Another study by Kuller et al. [61] also concluded that albumin could be a marker for coronary heart disease (CHD) as lower albumin could lead to persistent injury in arteries and the progression of atherosclerosis and thrombosis. Albuminuria is commonly found as a CVD risk factor in diabetic patients [62].
Table 10 summarizes the literature findings for the five potential biomarkers and their impacts on AMI patients. The findings from this study suggested that these five features could be used as potential biomarkers to predict the in-hospital mortality of AMI patients.
4.2 Advantages/strengths
Several strengths of this work as stated in the introduction are demonstrated in this section. Table 11 summarizes the comparison between some previous studies and this current study. In this work, we presented and compared the performance of different feature selection methods (filter, wrapper, embedded). In comparison, those previous studies either not included feature selection methods such as in Zhao et al. [67], or the models built were with more than or equal to 10 features as in Than et al. [68] and Ranga et al. [25]. Besides, there was no comparison between different types of feature selection methods (one type only). A recent study [69] includes a comparison between two wrapper methods (sequential floating forward selection and RFE), but that was a single method involved.
The most distinct finding of this study was the identification of a five-feature model, which achieved comparable results with the models that contained more features. From the medical point of view, fewer features for diagnosis or prognosis could reduce in cost and time of a patient, which indicates that fewer clinical and pathological tests are needed. Similarly, from the computational point of view, fewer features will effectively save the computational cost, and power and speed up the training time in building the classification models while increasing or retaining the model performance.
On top of that, those previous studies used AUROC instead of AUPRC as a model evaluation tool without considering the class imbalanced issue in their datasets [70]. Most CVD studies contained imbalanced datasets, yet AUROC or accuracy were chosen to evaluate their performance. On the other hand, this work utilized AUPRC as a top measurement to measure the model performance.
4.3 Challenges and limitations
There are several restrictions in this study. First, the number of samples is small, and the dataset consists of imbalanced classes. Hence, more validation works are needed to further confirm the reliability and viability of the proposed five biomarkers. Class imbalance is one of the most significant issues in machine learning. The trained models favoured performing poorly on the minority class when the dataset is imbalanced.
Secondly, the samples collected in this study were limited to only a single hospital (UMMC) compared to other studies, such as Than [68], which used data from nine centres. The richness of data and information would be higher from heterogeneous data of various centres. This would increase the potential of machine learning ability and robustness of the models, as well as increasing the chance of identifying better potential biomarkers.
Third, only one type of CVD, namely AMI, was involved in this study. Different types of AMI such as STEMI and NSTEMI as well as other CVDs such as coronary artery disease or peripheral arterial disease can be included.
This study also did not involve ECG findings due to the availability of data which is commonly included in AMI diagnosis as recommended in clinical practice guidelines [17, 71] nor any medical imaging data [8]. Features from imaging data like ECG findings are crucial in determining the type and location of AMI along with the risk of the patients.
5 Conclusion
An integrated model of feature selection and machine learning–based prognostic model had been developed. It was proven that the feature selection method did increase the performance of models as only the optimum features were selected. RF was the best classifier in all models with mean AUPRC > 0.8 (Full model = 0.8044; RFE-13 = 0.8260; RFI-13 = 0.8505; 5-feature model = 0.8462). The significant findings from this study are the identification of five clinicopathologic features for the in-hospital mortality classification of AMI patients namely cTnI, HDL cholesterol, HbA1c, anion gap, and albumin which were verified by the previous studies to be the significant risk stratification indicators to AMI/CVD. Hence, the combination of these five features could be used as potential biomarkers for the early detection and treatment of AMI. However, further research with larger and more diverse datasets is needed to validate the results and ensure generalizability to different populations. Then, the data could be classified further into different types of AMI such as STEMI and NSTEMI or other CVDs such as coronary artery disease or peripheral arterial disease. In the case where ECG data or other imaging data (cMRI) are to be included, the work could be expanded to multiclass classification rather than simple binary classification. In addition, a real-time data stream could be added to overcome data availability and improve accessibility apart from applying advanced technology, e.g., Internet of Things (IoT). Last but not least, future work opts to include and look into the prospects of different mortality rates among different ethnicities to compare the differences among them and identify potential disparities in AMI access and outcomes. Overall, this study presents a promising approach to the biomarker discovery of AMI using machine learning and feature selection methods.
References
Venkatason P et al (2019) In-hospital mortality of cardiogenic shock complicating ST-elevation myocardial infarction in Malaysia: a retrospective analysis of the Malaysian National Cardiovascular Database (NCVD) registry. BMJ Open 9(5):e025734
World Health Organization (WHO) (2021) Cardiovascular Disease. Available from: https://www.who.int/cardiovascular_diseases/en. Accessed 13 Oct 2021
Amir M, Mappiare M, Indra P (2017) The impact of cytochrome P450 2C19 polymorphism on cardiovascular events in indonesian patients with coronary artery disease. Clin Cardiol Cardiovasc Med 1:15–21
Ang CS, Chan KM (2016) A review of coronary artery disease research in Malaysia. Med J Malaysia 74:67–78
Institute for Health Metrics and Evaluation (IHME) (2020) GBD 2019 cause and risk summary: cardiovascular disease. Available from: https://www.healthdata.org/results/gbd_summaries/2019. Accessed 9 Apr 2022
Mahidin UDoSM (2020) Statistics on causes of deaths, Malaysia, 2020. Available from: https://www.dosm.gov.my/v1/index.php?r=column/cthemeByCat&cat=401&bul_id=QTU5T0dKQ1g4MHYxd3ZpMzhEMzdRdz09&menu_id=L0pheU43NWJwRWVSZklWdzQ4TlhUUT09. Accessed 13 Oct 2021
Mohamad BDoSM (2022) Statistics on causes of death, Malaysia, 2022. Available from: https://www.dosm.gov.my/portal-main/release-content/statistics-on-causes-of-death-malaysia-2022#:~:text=Ischaemic%20heart%20diseases%20was%20the,17%2C708%20and%2013%2C355%20deaths%2C%20respectively. Accessed 21 Feb 2023
MohdFaizal AS et al (2021) A review of risk prediction models in cardiovascular disease: conventional approach vs. artificial intelligent approach. Comput Methods Programs Biomed 207:106190
Antman EM et al (2000) The TIMI risk score for unstable angina/non-ST elevation MI: a method for prognostication and therapeutic decision making. JAMA 284(7):835–842
Mueller H, Rao A, Forman S (1987) Thrombolysis in myocardial infarction (TIMI): comparative studies of coronary reperfusion and systemic fibrinogenolysis with two forms of recombinant tissue-type plasminogen activator. J Am Coll Cardiol 10(2):479–490
Brindle P et al (2003) Predictive accuracy of the Framingham coronary risk score in British men: prospective cohort study. BMJ 327(7426):1267
Su TT et al (2015) Prediction of cardiovascular disease risk among low-income urban dwellers in Metropolitan Kuala Lumpur. Malaysia BioMed Res Int 2015:516984
Anand A et al (2020) Frailty assessment and risk prediction by GRACE score in older patients with acute myocardial infarction. BMC Geriatr 20:1–9
Hung J et al (2020) Performance of the GRACE 2.0 score in patients with type 1 and type 2 myocardial infarction. Eur Heart J 42(26):2552–2561
Backus B et al (2013) A prospective validation of the HEART score for chest pain patients at the emergency department. Int J Cardiol 168(3):2153–2158
Aydin S et al (2019) Biomarkers in acute myocardial infarction: current perspectives. Vasc Health Risk Manag 2019(15):1
Thygesen K et al (2018) Fourth universal definition of myocardial infarction. J Am Coll Cardiol 72(18):2231–2264
Martinez P et al (2019) Biomarkers in acute myocardial infarction diagnosis and prognosis. Arq Bras Cardiol 113:40–41
IBM Cloud Education (2020) Machine Learning. Available from: https://www.ibm.com/topics/machine-learning. Accessed 20 Apr 2022
Bansal A et al (2020) Machine learning techniques to predict in-hospital cardiovascular outcomes in elderly patients presenting with acute myocardial infarction. J Am College Cardiol 75(11_Supplement_1):360
Hazrani Abdul Halim M, SuhaylahYusoff Y, Yusuf MMD (2018) Predicting sudden deaths following myocardial infarction in Malaysia using machine learning classifiers. J Int J Eng Technol 7(4.15):3
Jaafar J et al (2013) Evaluation of machine learning techniques in predicting acute coronary syndrome outcome. Springer International Publishing, Cham
Li X et al (2017) Using machine learning models to predict in-hospital mortality for ST-elevation myocardial infarction patients. Stud Health Technol Inform 245:476–480
Mansoor H et al (2017) Risk prediction model for in-hospital mortality in women with ST-elevation myocardial infarction: a machine learning approach. Heart Lung 46(6):405–411
Ranga V, Rohila D (2018) Parametric analysis of heart attack prediction using machine learning techniques. Int J Grid Distrib Comput 11(4):37–48
Alaa AM et al (2019) Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK Biobank participants. PLoS One 14(5):e0213653
Sherazi SWA et al (2020) A machine learning–based 1-year mortality prediction model after hospital discharge for clinical patients with acute coronary syndrome. Health Informatics J 26(2):1289–1304
Aziz F et al (2021) Short- and long-term mortality prediction after an acute ST-elevation myocardial infarction (STEMI) in Asians: a machine learning approach. PLOS ONE 16(8):e0254894
Rengaraju U (2020) Handling missing values. Medium. Available from: https://medium.com/wids-mysore/handling-missing-values-82ce096c0cef. Accessed 30 Apr 2022
Charfaoui Y (2020) Hands-on with feature engineering techniques: imputing missing values. Available from: https://heartbeat.fritz.ai/hands-on-with-feature-engineering-techniques-imputing-missing-values-6c22b49d4060. Accessed 30 Apr 2022
Gogia N (2019) Why scaling is important in machine learning? Available from: https://medium.com/analytics-vidhya/why-scaling-is-important-in-machine-learning-aee5781d161a. Accessed 23 Feb 2023
Fernando J (2021) Correlation coefficient. Avaialble from: https://www.investopedia.com/terms/c/correlationcoefficient.asp. Accessed 23 Feb 2022
Nickolas S (2021) What do correlation coefficients positive, negative, and zero means?. Available from: https://www.investopedia.com/ask/answers/032515/what-does-it-mean-if-correlation-coefficient-positive-negative-or-zero.asp. Accessed 23 Feb 2022
Paul S (2020) Beginner’s guide to feature selection in Python. Available from: https://www.datacamp.com/tutorial/feature-selection-python. Accessed 31 Mar 2021
Płoński P (2020) Random forest feature importance computed in 3 ways with Python. Available from: https://mljar.com/blog/feature-importance-in-random-forest/. Accessed 28 Aug 2021
do Nascimento PM et al (2020) A decision tree to improve identification of pathogenic mutations in clinical practice. BMC Med Inform Decis Mak 20(1):52
Yang L et al (2020) Study of cardiovascular disease prediction model based on random forest in eastern China. Sci Rep 10(1):5245
Chen L et al (2020) Voice disorder identification by using Hilbert-Huang Transform (HHT) and K nearest neighbor (KNN). J Voice 35(6):932.e1–932.e11
Eetemadi A, Tagkopoulos I (2019) Genetic neural networks: an artificial neural network architecture for capturing gene expression relationships. Bioinformatics 35(13):2226–2234
Chen H, Chen L (2017) Support vector machine classification of drunk driving behaviour. Int J Environ Res Public Health 14(1):108
Wang Y et al (2019) Comparison of machine learning models and Framingham Risk Score for the prediction of the presence and severity of coronary artery diseases by using Gensini Score. Research Square. https://doi.org/10.21203/rs.2.12128/v1
Brownlee J (2020) How to calculate precision, recall, and F-measure for imbalanced classification. Available from: https://machinelearningmastery.com/precision-recall-and-f-measure-for-imbalanced-classification/. Accessed 5 July 2021
Brownlee J (2020) ROC curves and precision-recall curves for imbalanced classification. Available from: https://machinelearningmastery.com/roc-curves-and-precision-recall-curves-for-imbalanced-classification/. Accessed 25 July 2021
Fu GH, Yi LZ, Pan J (2019) Tuning model parameters in class-imbalanced learning with precision-recall curve. Biom J 61(3):652–664
Draelos R (2019) Measuring performance: AUPRC and average precision. Available from: https://glassboxmedicine.com/2019/03/02/measuring-performance-auprc/. Accessed 6 Feb 2020
Chopra S (2019) An introduction to building a classification model using random forests in Python. Available from: https://blogs.oracle.com/ai-and-datascience/post/an-introduction-to-building-a-classification-model-using-random-forests-in-python. Accessed 7 June 2020
Zhang Z (2014) Too much covariates in a multivariable model may cause the problem of overfitting. J Thorac Dis 6(9):E196–E197
Kumari B, Swarnkar T (2011) Filter versus wrapper feature subset selection in large dimensionality micro array: a review. (IJCSIT) Int J Comput Sci Inf Technol 2(3):1048–1053
Charfaoui Y(2020) Hands-on with feature selection techniques: wrapper methods. Available from: https://medium.com/@mxcsyounes/hands-on-with-feature-selection-techniques-wrapper-methods-5bb6d99b1274. Accessed 23 May 2020
Gupta A (2020) Feature selection techniques in machine learning. Available from: https://www.analyticsvidhya.com/blog/2020/10/feature-selection-techniques-in-machine-learning/. Accessed 17 May 2021
Wanamaker BL et al (2019) Relationship between troponin on presentation and in-hospital mortality in patients with ST-segment-elevation myocardial infarction undergoing primary percutaneous coronary intervention. J Am Heart Assoc 8(19):e013551
Matetzky S et al (2000) Elevated troponin I level on admission is associated with adverse outcome of primary angioplasty in acute myocardial infarction. Circulation 102(14):1611–1616
Rice MS, MacDonald DC (1999) Appropriate roles of cardiac troponins in evaluating patients with chest pain. J Am Board Fam Pract 12(3):214–218
Lee CH et al (2016) Roles of high-density lipoprotein cholesterol in patients with acute myocardial infarction. Medicine (Baltimore) 95(18):e3319
Salonen JT et al (1991) HDL, HDL2, and HDL3 subfractions, and the risk of acute myocardial infarction. A prospective population study in eastern Finnish men. Circulation 84(1):129–139
Salinero-Fort MA et al (2021) Cardiovascular risk factors associated with acute myocardial infarction and stroke in the MADIABETES cohort. Sci Rep 11(1):15245
Pan W et al (2019) Prognostic value of HbA1c for in-hospital and short-term mortality in patients with acute coronary syndrome: a systematic review and meta-analysis. Cardiovasc Diabetol 18(1):169
Sahu A, Cooper HA, Panza JA (2006) The initial anion gap is a predictor of mortality in acute myocardial infarction. Coron Artery Dis 17(5):409–412
Tang Y et al (2020) Serum anion gap is associated with all-cause mortality among critically ill patients with congestive heart failure. 2020: https://doi.org/10.1155/2020/8833637
Islam MS et al (2019) Serum albumin level and in-hospital outcome of patients with first attack acute myocardial infarction. Mymensingh Med J 28(4):744–751
Kuller LH et al (1991) The relation between serum albumin levels and risk of coronary heart disease in the multiple risk factor intervention trial. Am J Epidemiol 134(11):1266–1277
Rawshani A et al (2019) Relative prognostic importance and optimal levels of risk factors for mortality and cardiovascular outcomes in type 1 diabetes mellitus. Circulations 39(16):1900–1912
Beckerman JM (2020) FACC heart disease and lowering cholesterol. Available from: https://www.webmd.com/heart-disease/features/how-can-i-prevent-heart-disease. Accessed 17 May 2021
Khan HA, Alhomida AS, Sobki SH (2013) Lipid profile of patients with acute myocardial infarction and its correlation with systemic inflammation. Biomarker insights 8. https://doi.org/10.4137/BMI.S110
Hermanides RS et al (2020) Impact of elevated HbA1c on long-term mortality in patients presenting with acute myocardial infarction in daily clinical practice: insights from a ‘real world’ prospective registry of the Zwolle Myocardial Infarction Study Group. Eur Heart J Acute Cardiovasc Care 9(6):616–625
Yang SW et al (2017) The serum anion gap is associated with disease severity and all-cause mortality in coronary artery disease. J Geriatr Cardiol 14(6):392–400
Zhao J et al (2021) Optimized machine learning models to predict in-hospital mortality for patients with ST-segment elevation myocardial infarction. Ther Clin Risk Manag 2021(17):951–961
Than MP et al (2019) Machine learning to predict the likelihood of acute myocardial infarction. Circulation 140(11):899–909
Farah C, Adla YA, Awad M (2022) Can machine learning predict mortality in myocardial infarction patients within several hours of hospitalization? A comparative analysis. In 2022 IEEE 21st Mediterranean Electrotechnical Conference (MELECON) (pp. 1135–1140). IEEE
Ozenne B, Subtil F, Maucort-Boulch D (2015) The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J Clin Epidemiol 68(8):855–859
Knuuti J et al (2020) 2019 ESC guidelines for the diagnosis and management of chronic coronary syndromes. Eur Heart J 41(3):407
Funding
This work was supported by the University of Malaya Impact Oriented Interdisciplinary Research Grant under project numbers IIRG020B-2019 and IIRG020C-2019. The funders were not involved in the study design, the data collection and analysis, the decision to publish, or the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohd Faizal, A.S., Hon, W.Y., Thevarajah, T.M. et al. A biomarker discovery of acute myocardial infarction using feature selection and machine learning. Med Biol Eng Comput 61, 2527–2541 (2023). https://doi.org/10.1007/s11517-023-02841-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02841-y