Abstract
In plants, isoprene plays a dual role: (a) as thermo-protective agent proposed to prevent degradation of enzymes/membrane structures involved in photosynthesis, and (b) as reactive molecule reducing abiotic oxidative stress. The present work addresses the question whether suppression of isoprene emission interferes with genome wide transcription rates and metabolite fluxes in grey poplar (Populus x canescens) throughout the growing season. Gene expression and metabolite profiles of isoprene emitting wild type plants and RNAi-mediated non-isoprene emitting poplars were compared by using poplar Affymetrix microarrays and non-targeted FT-ICR-MS (Fourier transform ion cyclotron resonance mass spectrometry). We observed a transcriptional down-regulation of genes encoding enzymes of phenylpropanoid regulatory and biosynthetic pathways, as well as distinct metabolic down-regulation of condensed tannins and anthocyanins, in non-isoprene emitting genotypes during July, when high temperature and light intensities possibly caused transient drought stress, as indicated by stomatal closure. Under these conditions leaves of non-isoprene emitting plants accumulated hydrogen peroxide (H2O2), a signaling molecule in stress response and negative regulator of anthocyanin biosynthesis. The absence of isoprene emission under high temperature and light stress resulted transiently in a new chemo(pheno)type with suppressed production of phenolic compounds. This may compromise inducible defenses and may render non-isoprene emitting poplars more susceptible to environmental stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Isoprene (2-methyl 1,3-butadiene) accounts for one-third of biogenic volatile organic compound (BVOC) fluxes in the atmosphere. It influences atmospheric chemistry and contributes to the formation of photosmog and ozone (Chameides et al. 1988; Biesenthal et al. 1997) because of its reactivity with reactive oxygen/nitrogen species (ROS/RNS). Most isoprene-emitting plants are dicotyledonous tree species, many of which belong to the genus Populus (Kesselmeier and Staudt 1999). Isoprene is synthesized in the chloroplast by isoprene synthase (ISPS) from dimethylallyl diphosphate (DMADP). In chloroplasts, DMADP is synthesized through the 2-C-methylerythritol-4-phosphate (MEP)-pathway (Rohmer et al. 1993) and together with its isomer isopentenyl diphosphate (IPP) is the common precursor for all terpenes. Under unstressed conditions, recently fixed CO2 is the main carbon (C) source feeding photosynthetic intermediates into the MEP-pathway (e.g. Loreto and Sharkey 1993). However, alternative cytosolic C sources are used for isoprene formation when photosynthetic C assimilation becomes limited (Karl et al. 2002; Schnitzler et al. 2004; Brilli et al. 2007). In unstressed poplar leaves, 1–10% of recently assimilated C ends in isoprene synthesis (Sharkey and Yeh 2001), and even higher proportions have been recorded under stress (Teuber et al. 2008).
The observation that isoprene emission represents a significant loss of photosynthetic carbon led to the proposal that this compound plays important physiological and ecological roles in the protection of plants from environmental stresses. The main hypothesis, that isoprene leads to stabilization of chloroplast membranes during high temperature events (Sharkey and Singsaas 1995), has been validated by fumigating non-emitters with isoprene (Singsaas et al. 1997), and by inhibiting isoprene emission chemically (Velikova et al. 2005a; Sharkey et al. 2001). Isoprene helps leaves to tolerate transient high temperature episodes (Sharkey et al. 2001; Loivamäki et al. 2007a; Sasaki et al. 2007), but also allows leaves to cope with high levels of reactive oxygen species (ROS) (Loreto and Velikova 2001; Affek and Yakir 2002). Recently, plant transformations allowed induction of isoprene biosynthesis in Arabidopsis (Loivamäki et al. 2007a; Sasaki et al. 2007) and tobacco (Laothawornkitkul et al. 2008; Vickers et al. 2009) as well as its suppression in poplar (Behnke et al. 2007). All transgenic approaches confirmed enhanced tolerance of isoprene emitting plants to high temperature or oxidative stress. However, all studies demonstrated that modulation of isoprene production also influenced other non-target metabolic pathways. For instance, isoprene-emitting Arabidopsis had higher growth rates under moderate thermal stress (Sasaki et al. 2007; Loivamäki et al. 2007a). This effect was not primarily due to isoprene-mediated protection of photosynthesis, but was rather based on retained growth capability (Loivamäki et al. 2007a). Behnke et al. (2009) reported that the repression of isoprene biosynthesis in poplar results in an up-regulation of compensatory antioxidants. In both studies, the target effect was accompanied by unpredictable secondary effects, including widespread metabolic shifts or changes of developmental processes.
The present work addresses the effect of RNAi-mediated suppression of isoprene emission on the primary and secondary metabolism of grey poplar. We focused on the regulation of metabolic C fluxes in the MEP-pathway by using targeted analyses of pathway intermediates and end products, and by assessing gene expression and activities of involved enzymes. In addition, we carried out non-targeted integrated analyses of the metabolome and the transcriptome, and showed that down-regulation of the expression of ISPS in poplar resulted in a reduced accumulation of phenolic compounds in mid-summer, when air temperature and light intensities were high.
Materials and methods
Cultivation of transgenic and wild-type poplars
Wildtype (WT) and selected transgenic P. x canescens lines were amplified by micropropagation as described in Loivamäki et al. (2007b). Acclimation of plants and cultivation under non-sterile conditions was done as described in Behnke et al. (2007).
After acclimation, the plants were planted into 2.2 L pots with the same soil substrate and cultivated in the greenhouse for one growing season (March 2006–Oct 2006). Growth parameters were measured in weekly intervals and climate data (greenhouse inside air temperature (HP-100-A, Imko, Ettlingen, Germany) and global radiation (Pyranometer CM11, Kipp & Zonen, Delft, The Netherlands)) were recorded as 30 min intervals throughout the experimental period. The greenhouse was ventilated to maintain nearly ambient temperature conditions and plants were watered regularly.
Seasonal sampling was performed at four time points during the 2006 growing season: June 27th and 28th, July 25th–27th, September 4th–6th and October 12th, 13th and 16th. At the first sampling date in June the harvested leaves were on average 18 days old, and experienced a mean air temperature of 23°C (Supplemental Fig. 1 online). At the following sampling dates in July, September and October leaves were on average 15, 42 and 68 days old, and experienced mean temperatures of 25, 19 and 18°C, respectively. Overall, leaf development was fastest in July.
To avoid known diurnal influences on emission rates, gene expression and enzyme activities, fully mature leaves (leaf #9 and #10, counted from the apex) were sampled at noon (for details, see Loivamäki et al. 2007b). In all cases we selected 10 WT and 10 empty vector control (pBinAR) plants and a minimum of five plants from five independent transformation events (35S::PcISPS-RNAi lines: RA1/2/17/18/22). For details of the selected lines see Behnke et al. (2007). The complete set of plants was analyzed to screen for isoprene emission, DMADP content and photosynthetic pigment concentration. For additional analyses, enzyme activities and gene transcript levels, as well as other metabolites, a subset of selected lines (WT, C, RA1, (n = 6 each), RA17 (n = 5)) was used. Due to a limitation of available mature leaves (#9/#10) in June the number of replicates was reduced from 6 and 5 to 3 leaves. Sampling for microarray and non-target metabolomic analyses were performed in parallel to the sampling in July. We sampled nine leaves each of WT and RA1 plants (#9 or #10) and always pooled 3 leaves to obtain 3 biological replicates.
Determination of DMADP and photosynthetic pigments
All metabolic analyses were performed on leaf material shock-frozen in liquid N2 and stored at −80°C until use. Determination of DMADP was done following the standard protocol established by Brüggemann and Schnitzler (2002). The quantification of carotenoids, xanthophylls and chlorophylls was performed as described in Behnke et al. (2007).
Analysis of amino acid contents
Aliquots of 100 mg of leaf powder were transferred in a mixture of 1 ml methanol:chloroform (7:3, v/v) and 0.2 ml HEPES buffer (5 mM EGTA, 20 mM HEPES, 10 mM NaF, pH 7). Homogenates were incubated on ice for 30 min. Water-soluble amino compounds were extracted twice with 0.6 ml distilled water. The aqueous phases were combined and freeze-dried (Alpha 2–4; Christ, Osterode, Germany). The dried material was dissolved in 1 ml of 0.02 M HCl directly before analyses. For derivatization 70 μl borate buffer and 20 μl derivatization reagent were added (both AccQ Tag™ Ultra derivatization kit, Waters, Milford, USA) to 10 μl sample in certified glass vials. Samples were incubated for 1 min at room temperature and subsequently for 15 min at 55°C. Analysis of amino acids was performed with an ultra performance liquid chromatography (UPLC) amino acid analysis (AAA) application system (Waters, Milford, USA). Separation was performed on an AccQTag™ Ultra column (100 mm × ID 2.1, particle 1.7 μm) at flow rate of 0.7 ml min−1 and column temperature at 61°C. The detection was at 254 nm via a tunable ultraviolet (TUV) optical detector. Peaks were identified and quantified via standards. Norvalin was used as internal standard. Data were analyzed using Empower software version 2 (Waters, Milford, USA).
Analysis of total phenolics and condensed tannins
Analysis of total phenolics was performed according to Makkar et al. (1993) and condensed tannins according to Porter et al. (1986).
Quantification of pyruvate and phosphoenolpyruvate
For the quantification of phosphoenolpyruvate (PEP), lyophilized leaf material was extracted with 10% HClO4/5% PVPP according to Weidmann et al. (1990). Pyruvate was extracted with 0.1 M HCl according to Lowry and Passoneau (1972) with modifications according to Einig and Hampp (1990). Phosphoenolpyruvate was determined by luminescence using the ADP assay with 50 μM ADP instead of PEP as substrate for the pyruvate kinase reaction (Hampp 1984). Quantification of pyruvate was according to Lowry and Passoneau (1972).
Quantification of leaf abscisic acid concentrations
The concentrations of abscisic acid were measured with a Phytodetek ABA enzyme immunoassay test kit according to the manufacturer’s protocol (Agdia Inc., Elkhart, USA).
Stable isotope and elemental analyses
The δ13C and total C were analyzed with an EA-IRMS system. A total of 0.2 mg of freeze dried leaf material were transferred in tin capsules (IVA Analysentechnik, Meerbusch, Germany) and combusted in an elemental analyzer (Flash EA 1112, Thermo Fisher Scientific, Milan, Italy) with a Porapack QS 50/80 mesh GC column (Waters, Milford, USA) coupled to a continuous-flow isotope-ratio mass spectrometer (DeltaPlusXP, Thermo Fisher Scientific, Bremen, Germany). The δ13C values were expressed in delta notation with respect to Vienna Peedee Belemnite (VPDB). IAEA-CH-6 (sucrose with a δ13C value of −10.449‰VPDB; IAEA, Vienna, Austria) was used as internal standard for analysis.
Measurement of isoprene emission rates
Functional screening of isoprene emission was performed with a head-space analysis system using proton transfer reaction mass spectrometry (PTR-MS) as described in Cinege et al. (2009). To avoid diurnal influences on emission rates, gene expression and enzyme activities (for details, see Loivamäki et al. 2007b), two leaf discs from leaves #9 and #10 of ~0.5 cm diameter were taken at noon (Central European Time) on sampling days. The obtained emission rates were normalized to standard conditions (30°C and 1,000 μmol m−2 s−1 PPFD) according to Guenther et al. (1995).
Quantitative RT–PCR
Quantitative measurements of transcription rates of phosphoenolpyruvate carboxylase (PcPEPC, accession number FN356197), deoxyxylulose-5-phosphate synthase (PcDXS, FN356201), deoxyxylulose-5-phosphate reductoisomerase (PcDXR, AJ574852), isoprene synthase (PcISPS, AJ294819) and phytoene synthase (PcPSY, AJ889824) were performed as described by Mayrhofer et al. (2005). For quantitative PCR measurements, the following oligonucleotide primer sets were used, resulting in the indicated PCR product lengths: PcISPS, forward 5′-tttgcctactttgccgtggttcaaaac-3′ and reverse 5′-tcctcagaaatgccttttgtacgcatg-3′ (202 bp); PcDXR, forward 5′-ggaatagtaggttgcgcaggc-3′ and reverse 5′-gcatatgtcttttccagcttctattgc-3′ (66 bp); PcDXS, forward 5′-gctctgtccattgccaatctta-3′ and reverse 5′-tcttccttcgccgttttcc-3′ (202 bp); PcPSY, forward 5′-atgcatcacatatcacacccaaa-3′ and reverse 5′-ctcctagcatcttctccaacatctc-3′ (379 bp); PcPEPC, forward 5′-tcggcgacggaacaagttaa-3′ and reverse 5′-tgcgtaggatgagcagtcaaga-3′ (201 bp). For all genes we chose elongation factor Iβ (PcEF, FN356200) as a gene for normalization: forward 5′-ccatagccaactggagccaat-3′ and reverse 5′-gatgatgagacagacatggcg-3′ (101 bp).
The pyruvate kinase (PcPK, FN356202) gene and the pyruvate phosphate dikinase (PcPPDK, FN356198) gene were chosen for RT–PCR validation of microarray results using normalized RT–PCR results and rma-normalized array intensities: PcPK, forward 5′-cctcgagctacaatttggttgc-3′ and reverse 5′-caatttggttccatggcca-3′ (77 bp); PcPPDK, 5′-attgcatgcctcaagcttacg-3′ and 5′-tgcacctttaccagtacgcttc-3′ (150 bp).
Determination of enzyme activity
For analysis of phosphoenolpyruvate carboxylase (PEPC) activities, protein extracts were performed as described in Loivamäki et al. (2007a). PEPC activity was measured using a modified protocol of Rosenstiel et al. (2004). For optimization of the assay for PEPC, the following modifications were made: 75 μl of protein extracts were mixed with 10 μl of 50 mM NaHCO3, 5 μl of MDH (0.1 U μl−1), 5 μl of 40 mM PEP and 5 μl of 40 mM NADH. The samples were incubated at 35°C and absorbances at 340 nm were measured after 20 and 50 min with a Nanodrop spectrometer (ThermoFisher Scientific, Waltham, USA) in UV–VIS mode.
Determination of in vivo H2O2/ROS accumulation
For in vivo visualization of accumulation of H2O2 small leaf discs of leaf #8 (wild type and 35S::PcISPS-RNAi line RA1) were infiltrated with 2’,7’-dichlorofluorescein diacetate (H2-DCFDA, 25 μM in 50 mM potassium phosphate buffer pH 7.0) and afterwards floated in potassium phosphate buffer for dark incubation at room temperature for 30 min. After incubation excess dye was removed from the surface by rinsing the leaf discs three times with buffer. Then leaf discs were either immediately scanned for H2O2/ROS detection or incubated at 30°C and 1,000 μmol m−2 s−1 PPFD for 30 min and 60 min, respectively, in a water bath. H2O2/ROS detection was done with a confocal laser scanning microscope (Zeiss LSM 510, Carl Zeiss Micro Imaging GmbH, Göttingen, Germany) as described by Hutzler et al. (1998). For data evaluation, the ratio of green versus red raw fluorescence intensities of mesophyll cells was used as a relative measure for H2O2 accumulation.
FT-ICR-MS and metabolomics
High-resolution mass spectra for molecular formula assignment were acquired on a Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR-MS, APEX Qe, Bruker, Bremen, Germany) equipped with a 12-Tesla superconducting magnet and an Apollo II electrospray (ESI) source. The samples were diluted in methanol to a methanolic concentration of 70% to give highest ion density inside the electrospray, without eliminating the neutral metabolites which are highly water-soluble. Each sample was introduced into the ionization source at a flow rate of 2 μL min−1 by a microliter pump, with a nebulizer gas pressure of 20 psi and a drying gas pressure of 15 psi (heated to 200°C). The ionization source was run once in the ESI(−) negative operation mode and once in the ESI(+) positive operation mode to generate mono-charged ions.
Spectra were externally calibrated on clusters of arginine (10 ppmv in methanol); calibration errors in the relevant mass range were always below 0.05 ppm. The spectra were acquired with a time domain of 1 Megaword with a mass-to-charge ratio (m/z) range of 146–2,000. The time domain transient length used for these measurements gave a mass resolving power of more than 100,000 at m/z = 350 Da. The spectra were zero filled to a processing size of 2 Megawords. Before Fourier transformation of the time-domain transient, a sine apodization was performed. No fragmentation experiments were performed in this study. Thus, the whole mass range could be scanned and 300 scans were summed up in each acquisition. The ion accumulation time in the ion source was set to 0.1 s. FT-ICR ESI(−) and ESI(+) spectra were exported to peak lists at a signal to noise (S/N) ratio of two and corrected to reach neutral mass lists that were combined. From those lists, unique masses in either WT or RA1 plants, as well as masses significantly two times higher in one poplar type were selected (t-test, P < 0.05).
Metabolite identification and possible assignment of the m/z values was performed via the MassTRIX web site (http://mips.gsf.de/proj/mbx/~masstrix/; Suhre and Schmitt-Kopplin 2008) with the following parameter settings: ‘Scan mode neutral (input data is already corrected)’, ‘Max. error 5 ppm’, ‘Database KEGG with expanded lipids’, ‘Organism/Populus trichocarpa (pop)/’ restricting to only Populus trichocarpa annotated data. Identified compounds were automatically mapped to KEGG/pop pathways using KEGG/API (http://www.genome.jp/kegg/soap/) via MassTRIX. The list of compounds assigned to KEGG pathways was supplemented by differentially expressed enzymes, obtained by Affymetrix microarray analyses (see below) similar to Janz et al. (unpublished).
Microarray analysis
Total RNA from frozen poplar leaves (#9/#10) was isolated with an Aurum Total RNA Mini Kit (Biorad, Hercules, USA) and cleaned up with the Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocols. Subsequent steps (RNA quality control, RNA processing and hybridization) for expression analyses with Affymetrix GeneChip® Poplar Genome Array (http://www.affymetrix.com/products_services/arrays/specific/poplar.affx) were done by the Microarray Facility, Tübingen, Germany (http://www.microarray-facility.com/). Background correction and quantile normalization of the Affymetrix CEL output files was computed applying the rma algorithm (Irizarry et al. 2003) using the affy package obtained from the Bioconductor software project for R (http://www.r-project.org/index.html). Statistical testing for differentially expressed genes was performed with the sam function from the siggenes package (Tusher et al. 2001). Before evaluation using the sam function, a filtering process was performed as proposed by Falcon and Gentleman (2007). First, non-expressed genes were removed by calculating Affymetrix’ MAS 5.0 change calls using the mas5calls function from the Affymetrix package. Only genes that had at least two ‘present’ calls in any one of the two genotypes were considered. Secondly, gene annotation was carried out via the PopArray database (http://aspendb.uga.edu/poparray) and probe sets with no annotation were removed. In the sam analysis, Delta was set to get an FDR of 10%. The resulting gene list was filtered for duplicate genes. In case of two-one probe sets matches to the same gene, the probe set with a higher expression ratio was used for further analysis if both genes showed the same tendency in gene expression. Preferentially, functional annotation from PHYTOZOME Populus trichocarpa web site was used and supplementary annotation from PopArray database.
Annotation data of differentially expressed genes was also checked for enzyme commission numbers (EC no.) and available EC nos. were mapped to KEGG/pop pathways via the MassTRIX web site together with the list of differentially regulated metabolites.
Accession numbers
Sequence data from this article can be found in the EMBL/Genbank data libraries under the following accession numbers: FN356201 (PcDXS), AJ574852 (PcDXR), AJ294819 (PcISPS), AJ 889824 (PcPSY), FN356197 (PcPEPC), FN356202 (PcPK), FN356200 (PcEF Iβ), FN356198 (PcPPDK). The Affymetrix GenChip® Poplar Genome Array data can be found under http://www.ebi.ac.uk/microarray-as/aer/entry. (Experiment: E-MEXP-2120).
Results
Suppression of isoprene synthase gene expression by RNAi results in stable non-isoprene emitting poplars
Down-regulation of PcISPS expression by RNAi was very effective resulting in stable lines with almost no isoprene emission throughout the growing season (Fig. 1a). Wildtype (WT) and empty vector control plants (C lines) displayed similar isoprene emission rates, being maximal in June (approximately 30 nmol m−2 s−1) and decreasing during the vegetation period to approximately 18–20 nmol m−2 s−1 in July and September and finally to 8–10 nmol m−2 s−1 in October.
Isoprenoid pathway metabolite screening of P. x canescens plants grown one season in a greenhouse. Analyzed were wild type plants (WT,
 , n = 10) empty vector control (C,
, n = 10) empty vector control (C,
 , n = 5–10), PcISPS-RNAi lines (RA1/2/17/18/22,
, n = 5–10), PcISPS-RNAi lines (RA1/2/17/18/22,
 , n = 13–27). a Standard isoprene emission rates, b dimethylallyl diphosphate (DMADP) content, c total carotenoid content (sum of Lut, Nx, Vx, Ax, Zx β-Cars), d sum chlorophyll a and chlorophyll b contents. ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.s. not significant
, n = 13–27). a Standard isoprene emission rates, b dimethylallyl diphosphate (DMADP) content, c total carotenoid content (sum of Lut, Nx, Vx, Ax, Zx β-Cars), d sum chlorophyll a and chlorophyll b contents. ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.s. not significant
The transcriptional blockage of the isoprene emission ‘valve’ resulted in a drastic accumulation of DMADP, the direct metabolic precursor of isoprene, from June to October as compared to WT and C lines (Fig. 1b, P < 0.05). DMADP content of non-isoprene emitting RNAi (RA) lines was highest in June (1,200 nmol g dw−1) whereas DMADP content in WT and C was highest in October (88/67 nmol g dw−1).
Isoprene and carotenoid biosynthesis are closely linked through the common metabolic precursor DMADP. However, we observed no difference in carotenoid levels (Fig. 1c) between WT, C and RA lines, except in October when RA lines had higher carotenoid levels compared to C but not to WT plants (P < 0.05). Generally, in October leaf carotenoid concentrations were lower compared to the earlier dates in the season.
Analysis of chlorophyll contents (Fig. 1d) also revealed no difference between lines, except for October where chlorophyll content was higher in RA lines compared to WT and C (P < 0.05).
Suppression of isoprene emission has no effect on terpene pathway genes
Expression of terpene and carotenoid biosynthesis-related genes coding for the initial steps (PcDXS (1-deoxy-D-xylulose-5-phosphate synthase) and PcDXR (1-deoxy-D-xylulose-5-phosphate reductoisomerase)) of the MEP-pathway, the branching to isoprene biosynthesis (PcISPS, isoprene synthase), as well as the PcPSY (phytoene synthase) gene, a regulatory step of carotenoid biosynthesis were measured in WT, C and RA1/17 lines. Similar gene expression profiles were found for PcDXS, PcDXR, and PcPSY (Supplemental Fig. 2A–D, online). For all three genes there was no clear seasonal trend in transcript levels, except that in October transcript levels clearly increased. PcISPS gene expression, as expected, was always close to zero in the non-isoprene emitting lines. For PcDXS, PcDXR and PcPSY no difference in transcript levels was detectable between the different genotypes.
Since PEPC (phosphoenolpyruvate carboxylase) activity controls the availability of PEP, which after transfer to chloroplasts and conversion to Pyr feeds plastidic isoprenoid biosynthesis (Rosenstiel et al. 2004), we analyzed gene expression and activity of this enzyme. The PcPEPC gene was highly expressed in June in the leaves of all three genotypes, whereas it was almost undetectable in July, September and October (Supplemental Fig. 3E, online). In contrast to the gene transcript accumulation pattern, we detected equal PEPC activity for all analyzed genotypes in the first 3 months and a strong increase of activity in October (Supplemental Fig. 2F, online).
Absence of isoprene emission did not feed back to Pyr/PEP and amino acid metabolism
Availability of pyruvate (Pyr), one of the substrates of DXS which catalyzes the entry step of the MEP-pathway, is thought to play a key role in the regulation of isoprene emission. Therefore, we quantified the amount of Pyr and of its activated form PEP. Pyr and PEP contents were highly variable with values ranging from 65-360 nmol g dw−1 and 140-470 nmol g dw−1, respectively (Fig. 2a, b). Overall, a clear pattern regarding differences in these metabolites between non-isoprene emitting and WT/C lines was never apparent; nor was there any difference in the regulatory PEP/Pyr ratio, which varied from 0.4 to 3.3 (Fig. 2c).
a Pyruvate (Pyr), b phosphoenolpyruvate (PEP) concentrations and c PEP/Pyr ratio of P. x canescens wild type plants (WT,
 , n = 3), empty vector control (C,
, n = 3), empty vector control (C,
 , n = 3–6) and PcISPS-RNAi lines (RA1/17,
, n = 3–6) and PcISPS-RNAi lines (RA1/17,
 , n = 4–9) cultivated for one growing season in the greenhouse. ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.d. not determined
, n = 4–9) cultivated for one growing season in the greenhouse. ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.d. not determined
Analysis of the 20 proteinogenic amino acids (Supplemental Figure S3, online) also revealed that absence of isoprene emission does not influence pattern nor concentration of these metabolites. In all genotypes, the content of amino acids, especially for the group originating from α-ketoglutarate, increased continuously over the season.
ABA levels and water use efficiency are not affected in non-isoprene emitting lines
Abscisic acid (ABA) biosynthesis is linked to isoprenoid metabolism via availability of the xanthophyll violaxanthin, whose cleavage supplies the ABA precursor. Over the growing season, ABA concentrations ranged from 14 to 44 pmol mg fw−1 without significant differences between genotypes (Fig. 3a). For all genotypes, highest ABA contents in leaves were detected in July. In addition to ABA, we analyzed the 13C stable carbon isotope ratios, expressed as δ13C, in whole leaves as an indicator of water use efficiency (WUE) (Fig. 3b) over the leaf life span. No difference in δ13C between lines was detectable; however, the δ13C values inversely correlated to leaf ABA content (R² = 0.796, P < 0.001). In July, the high levels of ABA, in conjunction with the reduced detection of 13C, indicate enhanced environmental stress, probably elicited by the high temperatures and light intensities during this phase of the year (Supplemental Fig. 1, online).
a Abscisic acid (ABA) concentrations and b carbon isotope discrimination (δ13C) of P.xcanescens plants grown one season in a greenhouse. Analyzed were wild type plants (WT,
 , n = 6), empty vector control (C,
, n = 6), empty vector control (C,
 , n = 6), PcISPS-RNAi lines (RA1/17,
, n = 6), PcISPS-RNAi lines (RA1/17,
 , n = 11). The δ13C values were expressed in delta notation with respect to Vienna Peedee Belemnite (VPDB). ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.s. not significant
, n = 11). The δ13C values were expressed in delta notation with respect to Vienna Peedee Belemnite (VPDB). ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.s. not significant
Reduced condensed tannin concentrations in non-isoprene emitting lines during July
Total phenolic content and condensed tannins, an important subgroup of phenolic metabolites, were measured in WT/C and in 5 non-isoprene emitting lines (RA1/2/17/18/22), at all sampling dates (Fig. 4).
a Total phenolic content and b proanthocyanidin concentration in P. x canescens wild type (WT,
 , n = 6), empty vector control (C,
, n = 6), empty vector control (C,
 , n = 4–6), PcISPS-RNAi lines (RA1/2/17/18/22,
, n = 4–6), PcISPS-RNAi lines (RA1/2/17/18/22,
 , n = 5–23) grown one season in a greenhouse. ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.s. not significant, n.d. not determined
, n = 5–23) grown one season in a greenhouse. ANOVA was performed for each sampling date separately (SPSS, P > 0.05), error bars represent the standard error of mean. n.s. not significant, n.d. not determined
The total phenolic content ranged from 18 mg g fw−1 to 30 mg g fw−1 (Fig. 4a). In June, September and October, the total amount of phenolic compounds was similar in the different genotypes. In July, overlapping with the stress period indicated by ABA and δ13C, we observed increased concentrations of phenolic compounds in WT and C plants (up to 25–30 mg g fw−1) compared to the non-isoprene emitting lines.
A similar but more pronounced pattern was found for condensed tannins (Fig. 4b). In July samples, the condensed tannin levels in WT and C plants were highest compared to other time points in the season and differed significantly from the levels present in non-isoprene emitting plants.
Metabolomic and transcriptomic shifts in isoprene versus non-isoprene emitting leaves from July samples
In July, leaves were exposed to the highest temperature and light intensities (Supplemental Fig. 1, online), i.e. to conditions when isoprene is required for thermal and oxidative protection. These leaves were analyzed to identify transcriptomic and metabolomic differences between isoprene-emitting (WT) and one non-emitting line (RA1) at this time point (Supplemental Tables S1, S2 and S3, online). Overall, we observed transcriptional down-regulation of 260 genes with functional annotation, in diverse metabolic and regulatory pathways in RA1 leaves. In addition, we identified 11 metabolites with annotated structure less abundant in RA1 compared to WT leaves. Only 5 metabolites were more concentrated in RA1 (Supplemental Table S1, online) then in WT. The differentially accumulated metabolites were attributed to KEGG/pop pathways together with 42 differentially expressed genes with KEGG/pop annotation (EC no.).
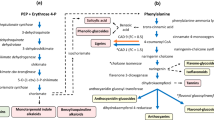
Supplemental Tables S1 and S2 summarize enzymes and metabolites with their corresponding KEGG/pop pathway assignments. Obviously, the most comprehensive changes were related to the metabolism of phenolic compounds including the KEGG/pop pathways ‘Phenylalanine, tyrosin and tryptophan biosynthesis’, ‘Phenylpropanoid biosynthesis’, ‘Flavonoid biosynthesis’ and ‘Flavone and flavonol biosynthesis’ (Fig. 5). Down-regulation of these pathways was mainly observed on the transcriptional level. Gene expression of the enzymes 3-deoxy-7-phosphoheptulonate synthase (DAHPS), 3-dehydroquinate dehydratase, shikimate dehydrogenase, 3-phosphoshikimate 1-carboxyvinyltransferase, chorismate synthase indicates a concerted down-regulation of biosynthesis of aromatic aa (Phe, Trp, Tyr) in the non-isoprene emitting line RA1. However, this down-regulation was not reflected by changed concentrations of aromatic amino acids (Fig. 2b). Down-regulation of gene expression proceeded to subsequent steps of biosynthesis of phenylpropanoids with relatively low transcript levels of genes coding for phenylalanine ammonia-lyase (PAL), 4-coumarate:CoA ligase (4CL), caffeoyl-CoA O-methyltransferase (COMT), cinnamyl-alcohol dehydrogenase (CAD), peroxidase, chalcone isomerase (CHI), and flavonol 3-O-glucosyltransferase (Fig. 5).
Analysis of pathways related to phenolics metabolism. Map displays selected steps from KEGG pathways pop00400 ‘Phenylalanine, tyrosin and tryptophan biosynthesis’, pop00940 ‘Phenylpropanoid biosynthesis’, pop00941 ‘Flavonoid biosynthesis’ and pop00944 ‘Flavone and flavonol biosynthesis’. Colors indicate significant expression, respective metabolite content ratios between P. x canescens PcISPS-RNAi line RA1 and wild type, blue indicates higher relative levels in wild type. Enzymes are given as EC numbers: 2.5.1.54, 3-deoxy-7-phosphoheptulonate synthase; 4.2.1.10, 3-dehydroquinate dehydratase; 1.1.1.25, shikimate dehydrogenase; 2.5.1.19, 3-phosphoshikimate 1-carboxyvinyltransferase; 4.2.3.5, chorismate synthase; 4.3.1.24, phenylalanine ammonia-lyase; 6.2.1.12, 4-coumarate:CoA ligase; 2.1.1.104, caffeoyl-CoA O-methyltransferase; 1.1.1.195, cinnamyl-alcohol dehydrogenase; 1.11.1.7, peroxidase; 5.5.1.6, chalcone isomerase; 2.4.1.91, flavonol 3-O-glucosyltransferase
The transcriptional down-regulation of functional genes involved in phenolic metabolism in line RA1 correlated with the lower abundance of only two compounds related to this pathway; C9H8O4 (possibly caffeate or 3-(4-hydroxyphenyl)pyruvate within this pathway), and C15H10O6 (kaempferol or luteolin). Nevertheless, concentrations of total phenolics and condensed tannins were lower in RA1 compared to WT leaves (Fig. 4).
Beside the down-regulation of phenolic metabolism in RA1 leaves, also pathways related to carbohydrate metabolism, in particular ‘Glycolysis/Gluconeogenesis’ and ‘Citrate cycle (TCA cycle)’, were down-regulated comprehensively in RA1 compared to WT leaves (Fig. 6). Expression of genes coding for several successive enzymatic steps of theses pathways were lower in RA1 plants (6-phosphofructokinase, fructose bisphosphate aldolase, 2-phospho-D-glycerate hydrolyase, pyruvate kinase, pyruvate dehydrogenase, dihydrolipoyl dehydrogenase, ATP citrate synthase, aconitate hydratase). Similar to the phenolic biosynthetic pathways only one KEGG/pop annotated compound assigned to these pathways (C6H14O12P2, fructose 1,6-bisphosphate) had lower abundance in RA1 compared to WT plants.
Analysis of pathways related to carbohydrate metabolism. Map displays selected steps from KEGG pathways pop00010 ‘Glycolysis/Gluconeogenesis’ and pop00020 ‘Citrate cycle (TCA cycle)’. Blue colors indicate higher relative levels of expression, respective metabolite content in P. x canescens wild type plants compared to PcISPS-RNAi line RA1. Enzymes are given as EC numbers: 2.7.1.11, 6-phosphofructokinase; 4.1.2.13, fructose bisphosphate aldolase; 4.2.1.11, 2-phospho-D-glycerate hydrolyase; 2.7.1.40, pyruvate kinase; 1.2.4.1, pyruvate dehydrogenase; 1.8.1.4, dihydrolipoyl dehydrogenase; 2.3.3.8, ATP citrate synthase
For validation of the microarray results by qRT-PCR, we chose the pyruvate kinase gene PcPK and the pyruvate phosphate dikinase gene PcPPDK (Supplemental Table S4, online). The qRT-PCR resulted in a fold change (log2(RA1 vs. WT)) of −0.52 for PcPK and of 0.72 for PcPPDK confirming the either down- or up-regulation of the two genes in RA1 plants as found with the microarray studies (Supplemental Table S4, online).
Transcript levels of genes that were not KEGG annotated confirm down-regulation of phenolic compounds in non-isoprene emitting plants
Several differentially regulated genes (201) could not be assigned to a KEGG/pop pathway. We grouped them according to a functional classification (Supplemental Table S3, online). Overall this list supports our KEGG-related interpretation of transcriptomic shifts in non-isoprene emitting line RA1 plants. Especially worth to emphasize are two additional putative genes coding for enzymes involved in glycolysis and the citrate cycle (2-phosphoglycerate kinase, phosphoglycerate mutase) as well as numerous genes coding for carbohydrate modifying enzymes (glycosyl transferases and glycoside hydrolases). Also the down-regulation of phenolic metabolism in RA1 plants is endorsed by lower expression of additional putative genes involved in biosynthesis of phenylpropanoids (anthocyanidin 3-O-glucosyltransferase, coumarate 3-hydroxylase, dihydroflavonol 4-reductase/flavanone 4-reductase).
Next to metabolic pathways, many differentially regulated transcripts play a role in regulatory pathways and code for signaling components. In particular, the lower transcript abundance of putative transcription factors known to regulate flavonoids and anthocyanin biosynthesis (orthologs of AtMYB4/AtMYB86, AtMYB58, AtTTG1) in RA1 plants indicated a transcriptional control of down-regulation of the phenolic biosynthesis in non-isoprene emitting leaves, which coincides with the reduced concentrations of total phenolic compounds and condensed tannins (Fig. 4).
We found the phosphoenolpyruvate carboxylase kinase (PEPC kinase) gene among the down-regulated PKs. PEPC kinase regulates the activity of PEPC and thus is centrally involved in the control of plant metabolism. However, this lower transcript abundance of PEPC kinase in RA1 plants did not result in lower PEPC activity in non-isoprene emitting lines relative to isoprene emitting lines as shown in Supplemental Fig. 2C.
Within the stress metabolism-associated transcripts, down-regulation of four genes encoding putative heat shock proteins (chaperones/co-chaperones) and several stress-induced proteins, disease resistance proteins and proteins playing a role in apoptosis/senescence was found in RA1 leaves, presumably indicating enhanced level of stress.
Pronounced accumulation of H2O2 in vivo under high temperature and light intensities in non-isoprene emitting leaves
We measured the formation of H2O2 in vivo in leaves of non-isoprene emitting poplars. Using DCFDA as molecular probe for H2O2 we visualized the accumulation of H2O2 in leaf discs from July samples that displayed the transcriptomic and metabolomic differences shown above. Depending on incubation time (0, 30, and 60 min at 1,000 μmol m−2 s−1 PPFD, 30°C), green fluorescence, and thus accumulation of H2O2, became visible in RA1 tissue, but not in WT (Fig. 7a). Prior to illumination the levels of H2O2 were low, and no difference in fluorescence between WT and RA1 was detectable, and also 60 min dark incubation did not result in H2O2 accumulation (Fig. 7b). Overall, upon exposure to light, a significant formation of H2O2 was observed only in the mesophyll layer of RA1 leaves (Fig. 7c).
H2O2 accumulation in P.
x
canescens wild type (WT) and PcISPS-RNAi line (RA1) during high light and temperature treatment (1,000 μmol m−2 s−1 PPFD and 30°C). Exemplary CLSM images of scanned area of leaf discs of illuminated WT and RA1 (a) and non-illuminated RA1 (b). Leaf discs were loaded with H2-DCFDA to assess ROS formation. The ratio of green versus red fluorescence intensities of total scanned area of CLSM images was used as marker for H2O2 accumulation (c). WT illuminated:  n = 5–8; RA1 illuminated:
n = 5–8; RA1 illuminated:
 n = 5–8; RA1 dark:
n = 5–8; RA1 dark:
 n = 3, error bars represent the standard error of mean
n = 3, error bars represent the standard error of mean
Discussion
Down-regulation of phenolic biosynthesis in non-isoprene emitting lines under high temperature
Our results clearly show a down-regulation of phenolic pathways in transgenic non-isoprene emitting grey poplars under high temperature and high light conditions, naturally associated with very high isoprene emission rates (e.g. Kesselmeier and Staudt 1999; Loreto et al. 2007). Our analysis demonstrated a concerted down-regulation of (a) shikimate pathway, (b) biosynthesis of aromatic amino acids, and (c) various branches of phenylpropanoid metabolism in non-isoprene emitting plants in July.
The down-regulation was most remarkable for the biosynthesis of phenolic compounds. The entry points of the phenolic pathways (DAHPS and PAL), as well as numerous subsequent genes, were found to be transcriptionally down-regulated in transgenic non-isoprene emitting grey poplars. However, at the metabolic levels only phenolic end products, total phenolics, anthocyanins (= flavonoid glycosides), and condensed tannins were less abundant in the transgenic, non-isoprene emitting plants, whereas the concentrations of intermediates of the shikimate pathway and aromatic amino acid biosynthesis were not affected. We assume that the reduced concentration of phenolic compounds reflects a response to environmental stress, namely high temperature, high light and water deficit. Indeed, differences in total phenolic and condensed tannin concentrations between genotypes were only present in July, under the highest temperature and light intensities experienced by the plants. Comparison of different poplar genotypes indicates a correlation between their phenolic content and drought tolerance (Street et al. 2006). Furthermore, poplars evolutionary adapted to desert climate contain, under non-stressed conditions, higher phenolic and tannin contents in their leaves than poplars evolved under temperate conditions (Janz et al., unpublished).
High temperature normally stimulates isoprene biosynthesis and emission, possibly to protect the photosynthetic machinery from thermal/oxidative damage caused by transient high temperature stress (Sharkey and Singsaas 1995; Singsaas and Sharkey 2000; Vickers et al. 2009). Such a thermoprotective function of isoprene has been successfully demonstrated with the same transgenic non-isoprene emitting grey poplar lines (Behnke et al. 2007, 2010) used in the present study. The cellular mechanism of thermal protection is still unclear, but the most accepted hypothesis is that isoprene is embedded in membranes and increases the stability of membranes and photosynthetic processes by preventing membrane lipid denaturation during oxidative stress (for overview see Loreto and Schnitzler 2010).
It is known that an induction of phenolic compound biosynthesis, mainly anthocyanins, occurs at low temperatures (Christie et al. 1994; Leyva et al. 1995; Bilger et al. 2007). A down-regulation of phenolic biosynthesis at high temperatures was also observed (Dela et al. 2003) confirming that temperature plays a key role in affecting anthocyanin accumulation in plant tissue. The mechanism reducing anthocyanin levels at high temperatures seems to be based on (a) the inhibition of mRNA transcription of biosynthetic genes and (b) the increased degradation of anthocyanins (Mori et al. 2007; Rowan et al. 2009). Rowan et al. (2009) investigated the environmental control of anthocyanin accumulation with a genetic approach using transgenic Arabidopsis thaliana plants constitutively over-expressing the PAP1 gene, a conserved MYB transcription factor. PAP1 is a key regulator of the phenylpropanoid biosynthesis and controls anthocyanin accumulation (Borevitz et al. 2000) in concert with other regulators associated in a heterotrimeric protein complex (MYB-bHLH-WD40; Ramsey and Glover 2005). Rowan et al. (2009) demonstrated that a complex MYB-TF system controls temperature sensitivity of anthocyanin biosynthetic genes involving activators (i.e. AtPAP1, AtTTG1, AtMYB4), which itself are regulated by repressors (i.e. AtMYB3, AtMYB6), that are up-regulated by high temperatures. The results of our microarray analysis pinpoint a comparable transcriptional control of phenolic compounds in the PcISPS-RNAi poplars at high temperatures. We found that orthologs of AtTTG1, coding for a WD40 protein, and AtMYB4 were down-regulated and therefore coincided with impaired anthocyanin biosynthesis at higher temperatures. Both transcription factors are possible activators of anthocyanin biosynthesis which are down-regulated at higher temperatures as demonstrated by Rowan et al. (2009). However, AtMYB4 was also shown to repress anthocyanin biosynthesis (Jin et al. 2000). Additionally, we found AtMYB58, a transcriptional activator of phenylpropanoid metabolism (Zhou et al. 2009), down-regulated in non-isoprene emitting RA1 leaves. Therefore, the down-regulated TFs may contribute to the comprehensive down-regulation of target genes (PAL, 4CL, CHI, LDOX, etc.), and loss of phenolic compounds, as shown in this study, and in Arabidopsis (Tohge et al. 2005; Rowan et al. 2009).
Vanderauwera et al. (2005) identified H2O2 to be a key molecule in the regulatory network responsible for impairment of anthocyanin biosynthesis. By comparing Arabidopsis WT and catalase (CAT)-deficient plants, a transcriptional cluster, which is induced in WT plants by high light intensities, was identified. This induction is impaired in the CAT-deficient plants due to elevated levels of H2O2. The transcriptional cluster includes MYB transcription factors (PAP1, PAP2) and the complete set of anthocyanin biosynthetic genes. In our study with poplar, the high temperature and light intensity in July resulted in an increased accumulation of phenolic compounds in WT and vector control plants, whereas in non-isoprene emitting genotypes this accumulation was inhibited. The difference of phenolic compounds might be linked to different H2O2 levels. Indeed, we observed an accumulation of H2O2 in leaf discs from non-isoprene emitting poplar leaves, but not in WT plants emitting high levels of isoprene, under combined high light and high temperature conditions. Thus, we hypothesize that H2O2 formation in the non-isoprene emitting plants during high light and high temperature episodes possibly mediates the down-regulation of the phenylpropanoid pathway.
The increased accumulation of H2O2 in non-isoprene emitting leaves of the PcISPS-RNAi line RA1 also confirms the proposed functions of isoprene as a ROS detoxifying agent (Affek and Yakir 2002; Loreto et al. 2001). Earlier work by Loreto and Velikova (2001) and Velikova et al. (2005b) clearly demonstrated that H2O2 and NO formation can be quenched in vivo by isoprene. We speculate that the environmental conditions in July stimulated increased H2O2 accumulation in the non-isoprene emitting plants and that changes in H2O2 concentration are involved in the signaling cascade triggering the down-regulation of transcription clusters including total phenolic compound biosynthesis, and particularly biosynthesis of condensed tannins and anthocyanins.
Implications on primary carbohydrate metabolism under high temperatures
A concerted regulation of glycolysis at the protein (Umeda and Uchimiya 1994; Centeno et al. 2008) and gene (Wasaki et al. 2003) level is believed to maintain a constant metabolic flux through the glycolytic pathway. We hypothesized that a metabolite-mediated regulation of gene expression is the basis of the observed down-regulation of glycolysis in the non-isoprene emitting plants. Metabolic repression of transcription is common, especially in carbohydrate metabolism (Sheen 1990; Yu 1999). There is evidence that excessive accumulation of P in phosphorylated intermediates can restrict glycolysis (Hohmann et al. 1996; Teusink et al. 1998). Therefore, a negative feedback loop on glycolysis due to the strong accumulation of P-rich DMADP might be possible. In our non-isoprene emitting poplar lines, the repression of isoprene biosynthesis led to a large accumulation of DMADP which might have been balanced by transcriptional control to reconstitute carbon allocation. The comprehensive alterations of carbohydrate metabolism at the transcriptional level were not paralleled by changes at the metabolic level, supporting adjustment of carbon sinks and sources.
Tight control of chloroplastic terpene biosynthesis
Here, we demonstrate that the substantial suppression of isoprene emission is stable during the growing season and results in an accumulation of excess DMADP in non-isoprene emitting poplars. Despite the accumulation of DMADP, no other measured parameter linked to plastidic terpene biosynthesis differed between isoprene emitting and non-isoprene emitting lines. In particular, neither gene expression of the initial steps (PcDXS and PcDXR) of the MEP-pathway, nor the PcPSY gene, a regulatory step of carotenoid biosynthesis, were different in isoprene emitting and non-emitting lines. Nevertheless, recent analysis of DXS activity in poplar leaf extracts (Ghirardo et al. 2010) showed lower DXS activity in non-isoprene emitting leaves compared to isoprene emitting ones. Thus, the absence of isoprene emission might negatively feedback on DXS activity due to the accumulation of DMADP. From feeding experiments with dideuterated 1-deoxy-D-xylulose (DOX-d2) (Wolfertz et al. 2004), bypassing the intrinsic DXP biosynthesis, a strong in vivo feedback regulation mediated by DMADP and/or other MEP-pathway intermediates is proposed.
As a consequence of yet not well understood regulatory steps, the abundance of chloroplastic carotenoids and ABA was not influenced by the transformation. The latter observation is of particular interest, as ABA biosynthesis occurs through the chloroplastic isoprenoid pathway, namely by cleavage of a violaxanthin intermediate (Schwartz et al. 2003). Evidence that a pool of foliar ABA could be controlled by the flow of carbon into the biosynthetic pathway of volatile isoprenoids was given from feeding experiments with fosmidomycin (FOS), a competitive inhibitor of DXR. Blockage of DXR activity with FOS resulted in a fast suppression of isoprene emission, accompanied by an increase in leaf transpiration and reduction of ABA concentration (Barta and Loreto 2006). However, such an effect was not present in non-isoprene emitting lines. It might be that ABA and isoprene biosynthesis in leaves exhibit only a weak tissue-specific overlap, since for Arabidopsis leaves it was shown (Endo et al. 2008) that ABA biosynthetic enzymes are localized most abundantly in vascular parenchyma cells at the boundary between xylem and phloem bundles while isoprene emission occurs in the mesophyll cells (Cinege et al. 2009). For this reason a metabolic competition between ABA and isoprene biosynthesis might be negligible. ABA was also of interest because it plays a general role in stress protection (Zeewart and Creelman 1988), and in particular regulates the plant water status (Acharya and Assmann 2009). Highest ABA concentrations in poplar were present in July, reflecting stomatal closure in response to drought stress since no direct correlation between ABA levels and temperature exists in poplar (Popko et al. 2010). Accordingly, the 13C enrichment in the leaf material indicated a reduced 13C discrimination of RubisCO, which can be attributed to stomatal closure and lower uptake of the heavier 13C isotope during photosynthetic carbon acquisition (Brugnoli and Farquhar 2000).
Extrachloroplastic metabolism may influence isoprene emission (Rosenstiel et al. 2004). Alternative, non-chloroplastic sources of carbon can contribute to isoprene biosynthesis (Karl et al. 2002; Schnitzler et al. 2004), mostly under stress conditions, when net assimilation is inhibited (Brilli et al. 2007). We did not observe any changes in PEP or Pyr concentrations, in the ratio between those metabolites, and in PEPC activity or gene expression, due to repression of isoprene biosynthesis, indicating that PEPC may not play an important regulatory role in the regulation of isoprene biosynthesis as earlier suggested (Rosenstiel et al. 2004; Loreto et al. 2007).
Conclusion
We analyzed transgenic non-isoprene emitting poplars during one growing season and assume that under natural conditions the quenching of hydrogen peroxide (and other reactive compounds) by isoprene not only prevents oxidative damage, but also influences intracellular stress perception and signaling cascades, which induce acclimation processes. This includes the down-regulation of phenolic compound biosynthesis under high temperature and light intensities as observed in this study. Our results indicate that the absence of isoprene emission in poplar under these circumstances generates a new transient chemo(pheno)type. Future experiments have to elucidate whether these changes impact plant sensitivity/susceptibility of non-isoprene emitting poplars towards abiotic (e.g. UV-B radiation, drought) and biotic (herbivory, fungal/microbial pathogens) stresses and overall biomass formation.
References
Acharya BR, Assmann SM (2009) Hormone interactions in stomatal function. Plant Mol Biol 69:451–462
Affek HP, Yakir D (2002) Protection by isoprene against singlet oxygen in leaves. Plant Physiol 129:269–277
Barta C, Loreto F (2006) The relationship between the methyl-erythritol phosphate pathway leading to emission of volatile isoprenoids and abscisic acid content in leaves. Plant Physiol 141:1676–1683
Behnke K, Ehlting B, Teuber M, Bauerfeind M, Louis S, Hänsch R, Polle A, Bohlmann J, Schnitzler JP (2007) Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J 51:485–499
Behnke K, Kleist E, Uerlings R, Wildt J, Rennenberg H, Schnitzler JP (2009). RNAi mediated suppression of isoprene biosynthesis impacts ozone tolerance. Tree Physiol 29:725–736
Behnke K, Loivamäki M, Zimmer I, Rennenberg H, Schnitzler JP, Louis S (2010) Isoprene emission protects photosynthesis in sunfleck exposed Grey poplar. Photosyn Res 104:5–17
Biesenthal TA, Wu Q, Shepson PB, Wiebe HA, Anlauf KG, MacKay GI (1997) A study of relationship between isoprene, its oxidation products, and ozone, in the lower Fraser Valley, BC. Atmos Environ 31:2049–2058
Bilger W, Rolland M, Nybakken L (2007) UV screening in higher plants induced by low temperature in the absence of UV-B radiation. Photochem Photobiol Sci 6:190–195
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Brilli F, Barta C, Fortunati A, Lerdau M, Loreto F, Centritto M (2007) Response of isoprene emission and carbon metabolism to drought in white poplar (Populus alba) saplings. New Phytol 175:244–254
Brüggemann N, Schnitzler JP (2002) Relationship of isopentenyl diphosphate (IDP) isomerase activity to isoprene emission of oak leaves. Tree Physiol 22:1011–1018
Brugnoli E, Farquhar GD (2000) Photosynthetic fractionation of carbon isotopes. In: Leegood RC, Sharkey TD, von Caemmerer S (eds) Photosynthesis: Physiology and Metabolism, Advances in Photosynthesis, vol 9. Kluwer Academic Publishers, Dordrecht, pp 399–434
Centeno DC, Oliver SN, Nunes-Nesi A, Geigenberger P, Machado DN, Ehlers Loureiro M, Silva MAP, Fernie AR (2008) Metabolic regulation of pathways of carbohydrate oxidation in potato (Solanum tuberosum) tubers. Physiol Plant 133:744–754
Chameides WL, Lindsay RW, Richardson J, Kiang CS (1988) The role of biogenic hydrocarbons in urban photochemical smog: Atlanta as a case study. Science 241:1473–1475
Christie PJ, Alfenito MR, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194:541–549
Cinege G, Louis S, Hänsch R, Schnitzler JP (2009) Regulation of isoprene synthase promoter by environmental and internal factors. Plant Mol Biol 69:593–604
Dela G, Or E, Ovadia R, Nissim-Levi A, Weiss D, Oren-Shamir M (2003) Changes in anthocyanin concentration and composition in ‘Jaguar’ rose flower due to transient high-temperature conditions. Plant Sci 164:333–340
Einig W, Hampp R (1990) Carbon partitioning in Norway spruce: amounts of fructose 2, 6-bisphosphate and of intermediates of starch/sucrose synthesis in relation to needle age and degree of needle loss. Trees 4:9–15
Endo A, Sawada Y, Takahashi H, Okamoto M, Ikegami K, Koiwai H, Seo M, Toyomasu T, Mitsuhashi W, Shinozaki K, Nakazono M, Kamiya Y, Koshiba T, Nambara E (2008) Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol 147:1984–1993
Falcon S, Gentleman R (2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23:257–258
Ghirardo A, Zimmer I, Brüggemann N, Schnitzler JP (2010) Analysis of 1-deoxy-d-xylulose 5-phosphate synthase activity in poplar leaves with isotope ratio mass spectrometry. Phytochemistry 71:918–922
Guenther A, Hewitt CN, Erickson D, Fall R, Geron C, Graedel T, Harley P, Klinger L, Lerdau M, McKay WA, Pierce T, Scholes B, Steinbrecher R, Tallamraju R, Taylor J, Zimmerman P (1995) A global model of natural volatile organic compound emissions. J Geophys Res 100:8873–8892
Hampp R (1984) Luminometric method. In: Bergmeyer J, Graßl M (eds) Methods in enzymatic analysis, vol 7. Verlag Chemie, Weinheim, pp 370–379
Hohmann S, Bell W, Neves AMJ, Valckx D, Thevelein JM (1996) Evidence for trehalose-6-phosphate-dependent and -independent mechanism in the control of sugar influx into yeast glycolysis. Mol Microbiol 20:981–991
Hutzler P, Fischbach R, Heller W, Jungblut TP, Reuber S, Schmitz R, Veit M, Weissenböck G, Schnitzler JP (1998) Tissue localisation of phenolic compounds in plants by confocal laser scanning microscopy. J Exp Bot 49:953–965
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Janz D, Behnke K, Schnitzler JP, Kanawati B, Schmitt-Kopplin P, Polle A (2010) Pathway analysis of the transcriptome and metabolome of a salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biol, under revision
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Karl T, Fall R, Rosenstiel TN, Prazeller P, Larsen B, Seufert G, Lindinger W (2002) On-line analyses of the 13CO2 labeling of leaf isoprene suggests multiple subcellular origins of isoprene precursors. Planta 215:894–905
Kesselmeier J, Staudt M (1999) Biogenic volatile organic compounds (VOC): an overview on emission, physiology, and ecology. J Atmos Chem 33:23–88
Laothawornkitkul J, Paul ND, Vickers CE, Possell M, Taylor JE, Mullineaux PM, Hewitt CN (2008) Isoprene emissions influence herbivore feeding decisions. Plant Cell Environ 31:1410–1415
Leyva A, Jarillo JA, Salinas J, Martinez-Zapater JM (1995) Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol 108:39–46
Loivamäki M, Gilmer F, Fischbach RJ, Sörgel C, Bachl A, Walter A, Schnitzler JP (2007a) Arabidopsis, a model to study biological functions of isoprene emission? Plant Physiol 144:1066–1078
Loivamäki M, Louis S, Cinege G, Zimmer I, Fischbach RJ, Schnitzler JP (2007b) Circadian rhythms of isoprene biosynthesis in Grey Poplar leaves. Plant Physiol 143:540–551
Loreto F, Schnitzler JP (2010) Abiotic stresses and induced BVOCs. Trend Plant Sci 15:154–166
Loreto F, Sharkey TD (1993) On the relationship between isoprene emission and photosynthetic metabolites under different environmental conditions. Planta 189:420–424
Loreto F, Velikova V (2001) Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol 127:1781–1787
Loreto F, Mannozzi M, Maris C, Nascetti P, Ferranti F, Pasqualini S (2001) Ozone quenching properties of isoprene and its antioxidant role in leaves. Plant Physiol 126:993–1000
Loreto F, Centritto M, Barta C, Calfapietra C, Fares S, Monson RK (2007) The relationship between isoprene emission rate and dark respiration rate in white poplar (Populus alba L.) leaves. Plant Cell Environ 30:662–669
Lowry OH, Passoneau JV (1972) A flexible system of enzymatic analysis. Academic Press, New York, p 291
Makkar H, Becker K, Abel H, Pawelzik E (1993) Gravimetric determination of tannins and their correlations with chemicals and protein precipitation methods. J Sci Food Agric 61:161–165
Mayrhofer S, Teuber M, Zimmer I, Louis S, Fischbach RJ, Schnitzler JP (2005) Diurnal and seasonal variation of isoprene biosynthesis-related genes in Grey poplar leaves. Plant Physiol 139:474–484
Mori K, Goto-Yamamoto N, Kitayama M, Hashizume K (2007) Loss of anthocyanins in red-wine grape under high temperature. J Exp Bot 58:1935–1945
Popko J, Hänsch R, Mendel RR, Polle A, Teichmann T (2010) The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol 12:242–258
Porter LJ, Hrstich LN, Chan BC (1986) The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochem 25:223–230
Ramsey NA, Glover BJ (2005) MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Rohmer M, Knanin M, Simonin P, Sutter P, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to the isopentenyl diphosphate. Biochem J 295:517–524
Rosenstiel TN, Ebbets AL, Khatri WC, Fall R, Monson RK (2004) Induction of poplar leaf nitrate reductase: a test of extrachloroplastic control of isoprene emission rate. Plant Biol 6:12–21
Rowan DD, Cao M, Lin-Wang K, Cooney JM, Jensen DJ, Austin PT, Hunt MB, Norling C, Hellens RP, Schaffer RJ, Allan AC (2009) Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol 182:102–115
Sasaki K, Saito T, Lämsä M, Oksman-Caldentey KM, Suzuki M, Ohyama K, Muranaka T, Ohara K, Yazaki K (2007) Plants utilize isoprene emission as a thermotolerance mechanism. Plant Cell Physiol 48:1254–1262
Schnitzler JP, Graus M, Kreuzwieser J, Heizmann U, Rennenberg H, Wisthaler A, Hansel A (2004) Contribution of different carbon sources to isoprene biosynthesis in poplar leaves. Plant Physiol 135:152–160
Schwartz S, Qin X, Zeewaart JAD (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131:1591–1601
Sharkey TD, Singsaas EL (1995) Why plants emit isoprene. Nature 374:769
Sharkey TD, Yeh S (2001) Isoprene emission from plants. Ann Rev Plant Phys Plant Mol Biol 52:407–436
Sharkey TD, Chen X, Yeh S (2001) Isoprene increases thermotolerance of fosmidomycin-fed leaves. Plant Physiol 125:2001–2006
Sheen J (1990) Metabolic repression of transcription in higher plants. Plant Cell 2:1027–1038
Singsaas EL, Sharkey TD (2000) Regulation of isoprene synthesis during high temperature stress. Plant Cell Environ 23:751–757
Singsaas EL, Lerdau M, Winter K, Sharkey TD (1997) Isoprene increases thermotolerance of isoprene emitting leaves. Plant Physiol 115:1413–1420
Street NR, Skogstrom O, Sjodin A, Tucker J, Rodriguez-Acosta M, Nilsson P, Jansson S, Taylor G (2006) The genetics and genomics of the drought response in Populus. Plant J 48:321–341
Suhre K, Schmitt-Kopplin P (2008) MassTRIX: mass translator into pathways. Nucleic Acids Res 36(Web Server issue): W481–484
Teuber M, Zimmer I, Kreuzwieser J, Ache P, Polle A, Rennenberg H, Schnitzler JP (2008) VOC emission of Grey poplar leaves as affected by salt stress and different N sources. Plant Biol 10:86–96
Teusink B, Walsh MC, van Dam K, Westerhoff HV (1998) The danger of metabolic pathways with turbo design. Trends Biochem Sci 23:162–169
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima JI, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Umeda M, Uchimiya H (1994) Differential transcript levels of genes associated with glycolysis and alcohol fermentation in rice plants (Oryza sativa L.) under submergence stress. Plant Physiol 106:1015–1022
Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139:806–821
Velikova V, Pinelli P, Loreto F (2005a) Consequences of inhibition of isoprene synthesis in Phragmites australis leaves exposed to elevated temperatures. Agric Ecosys Environ 106:209–217
Velikova V, Pinelli P, Pasqualini S, Reale L, Ferranti F, Loreto F (2005b) Isoprene decreases the concentration of nitric oxide in leaves exposed to elevated ozone. New Phytol 166:419–426
Vickers CE, Possell M, Cojocariu CI, Velikova VB, Laothawornkitkul J, Ryan A, Mullineaux PM, Hewitt CN (2009) Isoprene synthesis protects transgenic tobacco plants from oxidative stress. Plant Cell Environ 32:520–531
Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, Kishimoto N, Kikuchi S, Yamagishi M, Osaki M (2003) Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 26:1515–1523
Weidmann P, Einig W, Egger B, Hampp R (1990) Contents of ATP and ADP in needles of Norway spruce in relation to their development, age, and to symptoms of forest decline. Trees 4:68–74
Wolfertz M, Sharkey TD, Boland W, Kühnemann F (2004) Rapid regulation of the methylerythritol 4-phosphate pathway during isoprene synthesis. Plant Physiol 135:1939–1945
Yu SM (1999) Cellular and genetic responses of plants to sugar starvation. Plant Physiol 121:687–693
Zeewart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Ann Rev Plant Physiol Plant Mol Biol 39:439–473
Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21:248–266
Acknowledgments
We would like to thank S. Louis (IMK-IFU), W. Heller (BIOP Helmholtz Center Munich) and Kelly Vining (Oregon State University) for critical comments on the manuscript. The study was financially supported by the German Science Foundation (DFG; Schnitzler SCHN653/4, Hänsch HA3107/3, Rennenberg RE515/20, and Polle PO362/13) within the German joint research group ‘Poplar—A model to address tree-specific questions’, the Human Frontier Science Programme (HFSP, to J.-P. Schnitzler), and an exchange grant from the European Science Foundation (ESF) within the scientific program ‘VOCBAS’ (to C. Barta).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Behnke, K., Kaiser, A., Zimmer, I. et al. RNAi-mediated suppression of isoprene emission in poplar transiently impacts phenolic metabolism under high temperature and high light intensities: a transcriptomic and metabolomic analysis. Plant Mol Biol 74, 61–75 (2010). https://doi.org/10.1007/s11103-010-9654-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-010-9654-z