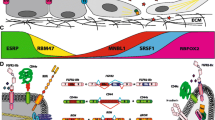
Abstract
Enhanced metastasis and disease recurrence accounts for the high mortality rates associated with cancer. The process of Epithelial-Mesenchymal Transition (EMT) contributes towards the augmentation of cancer invasiveness along with the gain of stem-like and the subsequent drug-resistant behavior. Apart from the well-established transcriptional regulation, EMT is also controlled post-transcriptionally by virtue of alternative splicing (AS). Numerous genes including Fibroblast Growth Factor receptor (FGFR) as well as CD44 are differentially spliced during this trans-differentiation process which, in turn, governs cancer progression. These splicing alterations are controlled by various splicing factors including ESRP, RBFOX2 as well as hnRNPs. Here, we have depicted the mechanisms governing the splice isoform switching of FGFR and CD44. Moreover, the role of the splice variants generated by AS of these gene transcripts in modulating the metastatic potential and stem-like/chemoresistant behavior of cancer cells has also been highlighted. Additionally, the involvement of splicing factors in regulating EMT/invasiveness along with drug-resistance as well as the metabolic properties of the cells has been emphasized. Tumorigenesis is accompanied by a remodeling of the cellular splicing profile generating diverse protein isoforms which, in turn, control the cancer-associated hallmarks. Therefore, we have also briefly discussed about a wide variety of genes which are differentially spliced in the tumor cells and promote cancer progression. We have also outlined different strategies for targeting the tumor-associated splicing events which have shown promising results and therefore this approach might be useful in developing therapies to reduce cancer aggressiveness in a more specific manner.


Similar content being viewed by others
Abbreviations
- AS:
-
Alternative splicing
- CD44:
-
Cluster of differentiation 44
- CSC:
-
Cancer stem cell
- EMT:
-
Epithelial-mesenchymal transition
- ESRP:
-
Epithelial splicing regulatory protein
- FGFR:
-
Fibroblast growth factor receptor
- hnRNP:
-
Heterogeneous nuclear ribonucleoprotein
- MET:
-
Mesenchymal-epithelial transition
References
Pucci B, Kasten M, Giordano A (2000) Cell cycle and apoptosis. Neoplasia 2:291–299. https://doi.org/10.1038/sj.neo.7900101
Cooper GM (2000) The cell: a molecular approach. The development and causes of cancer, 2nd edn. Sinauer Associates, Sunderland https://www.ncbi.nlm.nih.gov/books/NBK9963/
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. https://doi.org/10.1016/j.cell.2011.02.013
Koedoot E, Wolters L, van de Water B, Le Dévédec SE (2019) Splicing regulatory factors in breast cancer hallmarks and disease progression. Oncotarget 10:6021–6037. https://doi.org/10.18632/oncotarget.27215
Oltean S, Bates DO (2014) Hallmarks of alternative splicing in cancer. Oncogene 33:5311–5318. https://doi.org/10.1038/onc.2013.533
El Marabti E, Younis I (2018) The cancer spliceome: reprograming of alternative splicing in cancer. Front Mol Biosci 5:80. https://doi.org/10.3389/fmolb.2018.00080
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476. https://doi.org/10.1038/nature07509
Chen M, Manley JL (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10:741–754. https://doi.org/10.1038/nrm2777
Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI (2016) Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35:2413–2427. https://doi.org/10.1038/onc.2015.318
Escobar-Hoyos L, Knorr K, Abdel-Wahab O (2019) Aberrant RNA splicing in Cancer. Annu Rev Cancer Biol 3:167–185. https://doi.org/10.1146/annurev-cancerbio-030617-050407
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB (2011) An EMT–driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7:e1002218. https://doi.org/10.1371/journal.pgen.1002218
Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119:1420–1428. https://doi.org/10.1172/JCI39104
Smith BN, Bhowmick NA (2016) Role of EMT in metastasis and therapy resistance. J Clin Med 5:17. https://doi.org/10.3390/jcm5020017
Geiger TR, Peeper DS (2009) Metastasis mechanisms. Biochim Biophys Acta Rev Cancer 1796:293–308. https://doi.org/10.1016/j.bbcan.2009.07.006
Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP (2010) An ESRP-regulated splicing programme is abrogated during the epithelial–mesenchymal transition. EMBO J 29:3286–3300. https://doi.org/10.1038/emboj.2010.195
Reinke LM, Xu Y, Cheng C (2012) Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem 287:36435–36442. https://doi.org/10.1074/jbc.m112.397125
Wu HT, Zhong HT, Li GW, Shen JX, Ye QQ, Zhang ML, Liu J (2020) Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J Transl Med 18:51. https://doi.org/10.1186/s12967-020-02240-z
Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, Rousset F (2013) RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol 33:396–405. https://doi.org/10.1128/MCB.01174-12
Aponte PM, Caicedo A (2017) Stemness in cancer: stem cells, cancer stem cells, and their microenvironment. Stem Cells Int 2017:5619472. https://doi.org/10.1155/2017/5619472
Bhattacharya R, Mitra T, Ray Chaudhuri S, Roy SS (2018) Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J Cell Biochem 119:3373–3383. https://doi.org/10.1002/jcb.26504
Zhou P, Li B, Liu F, Zhang M, Wang Q, Liu Y, Yao Y, Li D (2017) The epithelial to mesenchymal transition (EMT) and cancer stem cells: implication for treatment resistance in pancreatic cancer. Mol Cancer 16:52. https://doi.org/10.1186/s12943-017-0624-9
Mitra T, Prasad P, Mukherjee P, Chaudhuri SR, Chatterji U, Roy SS (2018) Stemness and chemoresistance are imparted to the OC cells through TGFβ1 driven EMT. J Cell Biochem 119:5775–5787. https://doi.org/10.1002/jcb.26753
Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, Bapat SA (2009) Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells 27:2059–2068. https://doi.org/10.1002/stem.154
Wellner U, Brabletz T, Keck T (2010) ZEB1 in pancreatic cancer. Cancers (Basel) 2:1617–1628. https://doi.org/10.3390/cancers2031617
Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, Elela SA, Roux P, Lemaitre JM, Tazi J (2013) MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun 4:2480. https://doi.org/10.1038/ncomms3480
Pradella D, Naro C, Sette C, Ghigna C (2017) EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 16:8. https://doi.org/10.1186/s12943-016-0579-2
Begicevic RR, Falasca M (2017) ABC transporters in cancer stem cells: beyond chemoresistance. Int J Mol Sci 18:2362. https://doi.org/10.3390/ijms18112362
Yuan S, Tao F, Zhang X, Zhang Y, Sun X, Wu D (2020) Role of Wnt/β-catenin signaling in the Chemoresistance modulation of colorectal Cancer. Biomed Res Int 2020:9390878. https://doi.org/10.1155/2020/9390878
Banerjee AK, Bhattacharya R, Mal C (2020) HMG2D: a tool to identify miRNAs/drugs/genes associated with diseases like cancers. Meta Gene 24:100699. https://doi.org/10.1016/j.mgene.2020.100699
Wang BD, Lee NH (2018) Aberrant RNA splicing in cancer and drug resistance. Cancers (Basel) 10:458. https://doi.org/10.3390/cancers10110458
Siegfried Z, Karni R (2018) The role of alternative splicing in cancer drug resistance. Curr Opin Genet Dev 48:16–21. https://doi.org/10.1016/j.gde.2017.10.001
Yang Q, Zhao J, Zhang W, Chen D, Wang Y (2019) Aberrant alternative splicing in breast cancer. J Mol Cell Biol 11:920–929. https://doi.org/10.1093/jmcb/mjz033
Bhattacharya R, Ray Chaudhuri S, Roy SS (2018) FGF9-induced ovarian cancer cell invasion involves VEGF-A/VEGFR2 augmentation by virtue of ETS1 upregulation and metabolic reprogramming. J Cell Biochem 119:8174–8189. https://doi.org/10.1002/jcb.26820
Mitra T, Bhattacharya R (2020) Phytochemicals modulate cancer aggressiveness: a review depicting the anticancer efficacy of dietary polyphenols and their combinations. J Cell Physiol 235:7645–8863. https://doi.org/10.1002/jcp.29703
Bielli P, Pagliarini V, Pieraccioli M, Caggiano C, Sette C (2019) Splicing dysregulation as oncogenic driver and passenger factor in brain tumors. Cells 9:10. https://doi.org/10.3390/cells9010010
Turner N, Grose R (2010) Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10:16–129. https://doi.org/10.1038/nrc2780
Rebscher N, Deichmann C, Sudhop S, Fritzenwanker JH, Green S, Hassel M (2009) Conserved intron positions in FGFR genes reflect the modular structure of FGFR and reveal stepwise addition of domains to an already complex ancestral FGFR. Dev Genes Evol 219:455–468. https://doi.org/10.1007/s00427-009-0309-5
Holzmann K, Grunt T, Heinzle C, Sampl S, Steinhoff H, Reichmann N, Kleiter M, Hauck M, Marian B (2012) Alternative splicing of fibroblast growth factor receptor IgIII loops in cancer. J Nucleic Acids 2012:950508. https://doi.org/10.1155/2012/950508
Acevedo VD, Ittmann M, Spencer DM (2009) Paths of FGFR-driven tumorigenesis. Cell Cycle 8:580–588. https://doi.org/10.4161/cc.8.4.7657
Matsuda Y, Ueda J, Ishiwata T (2012) Fibroblast growth factor receptor 2: expression, roles, and potential as a novel molecular target for colorectal cancer. Pathol Res Int 2012:574768. https://doi.org/10.1155/2012/574768
Oltean S, Sorg BS, Albrecht T, Bonano VI, Brazas RM, Dewhirst MW, Garcia-Blanco MA (2006) Alternative inclusion of fibroblast growth factor receptor 2 exon IIIc in dunning prostate tumors reveals unexpected epithelial mesenchymal plasticity. Proc Natl Acad Sci U S A 103:14116–14121. https://doi.org/10.1073/pnas.0603090103
Amann T, Bataille F, Spruss T, Dettmer K, Wild P, Liedtke C, Mühlbauer M, Kiefer P, Oefner PJ, Trautwein C, Bosserhoff AK (2010) Reduced expression of fibroblast growth factor receptor 2IIIb in hepatocellular carcinoma induces a more aggressive growth. Am J Pathol 176:1433–1442. https://doi.org/10.2353/ajpath.2010.090356
Zhao Q, Caballero OL, Davis ID, Jonasch E, Tamboli P, Yung WA, Weinstein JN, Shaw K, Strausberg RL, Yao J (2013) Tumor-specific isoform switch of the fibroblast growth factor receptor 2 underlies the mesenchymal and malignant phenotypes of clear cell renal cell carcinomas. Clin Cancer Res 19:2460–2472. https://doi.org/10.1158/1078-0432.CCR-12-3708
Ishiwata T (2018) Role of fibroblast growth factor receptor-2 splicing in normal and cancer cells. Front Biosci (Landmark Ed) 23:626–639. https://doi.org/10.2741/4609
Ueda J, Matsuda Y, Yamahatsu K, Uchida E, Naito Z, Korc M, Ishiwata T (2014) Epithelial splicing regulatory protein 1 is a favorable prognostic factor in pancreatic cancer that attenuates pancreatic metastases. Oncogene 33:4485–4495. https://doi.org/10.1038/onc.2013.392
Ranieri D, Belleudi F, Magenta A, Torrisi MR (2015) HPV16 E5 expression induces switching from FGFR2b to FGFR2c and epithelial-mesenchymal transition. Int J Cancer 137:61–72. https://doi.org/10.1002/ijc.29373
Teles SP, Oliveira P, Ferreira M, Carvalho J, Ferreira P, Oliveira C (2020) Integrated analysis of structural variation and RNA expression of FGFR2 and its splicing modulator ESRP1 highlight the ESRP1amp-FGFR2norm-FGFR2-IIIchigh Axis in diffuse gastric Cancer. Cancers 12:70. https://doi.org/10.3390/cancers12010070
Osada AH, Endo K, Kimura Y, Sakamoto K, Nakamura R, Sakamoto K, Ueki K, Yoshizawa K, Miyazawa K, Saitoh M (2019) Addiction of mesenchymal phenotypes on the FGF/FGFR axis in oral squamous cell carcinoma cells. PLoS One 14:e0217451. https://doi.org/10.1371/journal.pone.0217451
Hopkins A, Coatham ML, Berry FB (2017) FOXC1 regulates FGFR1 isoform switching to promote invasion following TGFβ-induced EMT. Mol Cancer Res 15:1341–1353. https://doi.org/10.1158/1541-7786.MCR-17-0185
Sonvilla G, Allerstorfer S, Heinzle C, Stättner S, Karner J, Klimpfinger M, Wrba F, Fischer H, Gauglhofer C, Spiegl-Kreinecker S, Grasl-Kraupp B, Holzmann K, Grusch M, Berger W, Marian B (2010) Fibroblast growth factor receptor 3-IIIc mediates colorectal cancer growth and migration. Br J Cancer 102:1145–1156. https://doi.org/10.1038/sj.bjc.6605596
Qian X, Anzovino A, Kim S, Suyama K, Yao J, Hulit J, Agiostratidou G, Chandiramani N, McDaid HM, Nagi C, Cohen HW, Phillips GR, Norton L, Hazan RB (2014) N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene 33:3411–3421. https://doi.org/10.1038/onc.2013.310
Maehara O, Suda G, Natsuizaka M, Ohnishi S, Komatsu Y, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K, Shimazaki T, Kimura M, Asano A, Fujimoto Y, Ohashi S, Kagawa S, Kinugasa H, Naganuma S, Whelan KA, Nakagawa H, Nakagawa K, Takeda H, Sakamoto N (2017) Fibroblast growth factor-2–mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis 38:1073–1083. https://doi.org/10.1093/carcin/bgx095
Saito S, Morishima K, Ui T, Hoshino H, Matsubara D, Ishikawa S, Aburatani H, Fukayama M, Hosoya Y, Sata N, Lefor AK (2015) The role of HGF/MET and FGF/FGFR in fibroblast-derived growth stimulation and lapatinib-resistance of esophageal squamous cell carcinoma. BMC Cancer 15:82. https://doi.org/10.1186/s12885-015-1065-8
Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, Helfrich BA, Bunn PA, Heasley LE (2009) Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol 75:196–207. https://doi.org/10.1124/mol.108.049544
Liu J, Chen G, Liu Z, Liu S, Cai Z, You P, Ke Y, Lai L, Huang Y, Gao H, Zhao L, Pelicano H, Huang P, McKeehan WL, Wu CL, Wang C, Zhong W, Wang F (2018) Aberrant FGFR tyrosine kinase signaling enhances the Warburg effect by reprogramming LDH isoform expression and activity in prostate cancer. Cancer Res 78:4459–4470. https://doi.org/10.1158/0008-5472.CAN-17-3226
Xu M, Chen S, Yang W, Cheng X, Ye Y, Mao J, Wu X, Huang L, Ji J (2018) FGFR4 links glucose metabolism and chemotherapy resistance in breast cancer. Cell Physiol Biochem 47:151–160. https://doi.org/10.1159/000489759
Chen C, Zhao S, Karnad A, Freeman JW (2018) The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 11:64. https://doi.org/10.1186/s13045-018-0605-5
Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C (2011) CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Inv 121:1064–1074. https://doi.org/10.1172/JCI44540
Yin T, Wang G, He S, Liu Q, Sun J, Wang Y (2016) Human cancer cells with stem cell-like phenotype exhibit enhanced sensitivity to the cytotoxicity of IL-2 and IL-15 activated natural killer cells. Cell Immunol 300:41–45. https://doi.org/10.1016/j.cellimm.2015.11.009
Zhao S, Chen C, Chang K, Karnad A, Jagirdar J, Kumar AP, Freeman JW (2016) CD44 expression level and isoform contributes to pancreatic Cancer cell plasticity, invasiveness, and response to therapy. Clin Cancer Res 22:5592–5604. https://doi.org/10.1158/1078-0432.CCR-15-3115
Lin J, Ding D (2017) The prognostic role of the cancer stem cell marker CD44 in ovarian cancer: a meta-analysis. Cancer Cell Int 17:1–11. https://doi.org/10.1186/s12935-016-0376-4
Zöller M (2011) CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 11:254–267. https://doi.org/10.1038/nrc3023
Skandalis SS, Karalis TT, Chatzopoulos A, Karamanos NK (2019) Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal 63:109377. https://doi.org/10.1016/j.cellsig.2019.109377
Bourguignon LYW (2019) Matrix Hyaluronan-CD44 interaction activates MicroRNA and LncRNA signaling associated with Chemoresistance, invasion, and tumor progression. Front Oncol 9:492. https://doi.org/10.3389/fonc.2019.00492
Xu H, Wu K, Tian Y, Liu Q, Han N, Yuan X, Zhang L, Wu GS, Wu K (2016) CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int J Oncol 49:1343–1350. https://doi.org/10.3892/ijo.2016.3639
Chen Q, Gu M, Cai ZK, Zhao H, Sun SC, Liu C, Zhan M, Chen YB, Wang Z (2020) TGF β1 promotes epithelial to mesenchymal transition and stemness of prostate cancer cells by inducing PCBP1 degradation and alternative splicing of CD44. Cell Mol Life Sci (Epub ahead of print). https://doi.org/10.1007/s00018-020-03544-5
Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, Girard L, Behrens C, Wistuba II, Gazdar AF, Hayward NK, Minna JD (2016) ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest 126:3219–3235. https://doi.org/10.1172/JCI76725
Inoue K, Fry EA (2015) Aberrant splicing of estrogen receptor, HER2, and CD44 genes in breast cancer. Genet Epigenet 7:19–32. https://doi.org/10.4137/GEG.S35500
Miwa T, Nagata T, Kojima H, Sekine S, Okumura T (2017) Isoform switch of CD44 induces different chemotactic and tumorigenic ability in gallbladder cancer. Int J Oncol 51:771–780. https://doi.org/10.3892/ijo.2017.4063
Li L, Qi L, Qu T, Liu C, Cao L, Huang Q, Song W, Yang L, Qi H, Wang Y, Gao B, Guo Y, Sun B, Meng B, Zhang B, Cao W (2018) Epithelial splicing regulatory protein 1 inhibits the invasion and metastasis of lung adenocarcinoma. Am J Pathol 188:1882–1894. https://doi.org/10.1016/j.ajpath.2018.04.012
Zhang FL, Cao JL, Xie HY, Sun R, Yang LF, Shao ZM, Li DQ (2018) Cancer-associated MORC2-mutant M276I regulates an hnRNPM-mediated CD44 splicing switch to promote invasion and metastasis in triple-negative breast cancer. Cancer Res 78:5780–5792. https://doi.org/10.1158/0008-5472.CAN-17-1394
Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, Wellner U, Hopt UT, Brummer T, Brabletz S, Brabletz T, Stemmler MP (2015) A self-enforcing CD 44s/ZEB 1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer 137:2566–2577. https://doi.org/10.1002/ijc.29642
Miyazaki H, Takahashi RU, Prieto-Vila M, Kawamura Y, Kondo S, Shirota T, Ochiya T (2018) CD44 exerts a functional role during EMT induction in cisplatin-resistant head and neck cancer cells. Oncotarget 9:10029–10041. https://doi.org/10.18632/oncotarget.24252
Tsubouchi K, Minami K, Hayashi N, Yokoyama Y, Mori S, Yamamoto H, Koizumi M (2017) The CD44 standard isoform contributes to radioresistance of pancreatic cancer cells. J Radiat Res 58:816–826. https://doi.org/10.1093/jrr/rrx033
Biddle A, Gammon L, Fazil B, Mackenzie IC (2013) CD44 staining of cancer stem-like cells is influenced by down-regulation of CD44 variant isoforms and up-regulation of the standard CD44 isoform in the population of cells that have undergone epithelial-to-mesenchymal transition. PLoS One 8:e57314. https://doi.org/10.1371/journal.pone.0057314
Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, Deng Y, Hu X, Zhang J, Gao XD, Kang Y, Mercurio AM, Goel HL, Cheng C (2019) CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev 33:166–179. https://doi.org/10.1101/gad.319889.118
Suwannakul N, Ma N, Thanan R, Pinlaor S, Ungarreevittaya P, Midorikawa K, Hiraku Y, Oikawa S, Kawanishi S, Murata M (2018) Overexpression of CD44 variant 9: a novel cancer stem cell marker in human cholangiocarcinoma in relation to inflammation. Mediat Inflamm 2018:4867234. https://doi.org/10.1155/2018/4867234
Kiuchi S, Ikeshita S, Miyatake Y, Kasahara M (2015) Pancreatic cancer cells express CD44 variant 9 and multidrug resistance protein 1 during mitosis. Exp Mol Pathol 98:41–46. https://doi.org/10.1016/j.yexmp.2014.12.001
Omran OM, Ata HS (2012) CD44s and CD44v6 in diagnosis and prognosis of human bladder cancer. Ultrastruct Pathol 36:145–152. https://doi.org/10.3109/01913123.2011.651522
Wang Z, Tang Y, Xie L, Huang A, Xue C, Gu Z, Wang K, Zong S (2019) The prognostic and clinical value of CD44 in colorectal Cancer: a Meta-analysis. Front Oncol 9:309. https://doi.org/10.3389/fonc.2019.00309
Tjhay F, Motohara T, Tayama S, Narantuya D, Fujimoto K, Guo J, Sakaguchi I, Honda R, Tashiro H, Katabuchi H (2015) CD 44 variant 6 is correlated with peritoneal dissemination and poor prognosis in patients with advanced epithelial ovarian cancer. Cancer Sci 106:1421–1428. https://doi.org/10.1111/cas.12765
Chen L, Fu C, Zhang Q, He C, Zhang F, Wei Q (2020) The role of CD44 in pathological angiogenesis. FASEB J 34:13125–13139. https://doi.org/10.1096/fj.202000380RR
Papadaki C, Manolakou S, Lagoudaki E, Pontikakis S, Ierodiakonou D, Vogiatzoglou K, Messaritakis I, Trypaki M, Giannikaki L, Sfakianaki M, Kalykaki A, Mavroudis D, Tzardi M, Souglakos J (2020) Correlation of PKM2 and CD44 protein expression with poor prognosis in platinum-treated epithelial ovarian Cancer: a retrospective study. Cancers 12:1013. https://doi.org/10.3390/cancers12041013
Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, Sugihar E, Onishi N, Yamamoto T, Yanagawa H, Suematsu M, Saya H (2012) Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer Res 72:1438–1448. https://doi.org/10.1158/0008-5472.CAN-11-3024
Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J (2014) The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 33:1082–1092. https://doi.org/10.1038/onc.2013.50
Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK (2019) The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog 58:196–205. https://doi.org/10.1002/mc.22919
Wen J, Toomer KH, Chen Z, Cai X (2015) Genome-wide analysis of alternative transcripts in human breast cancer. Breast Cancer Res Treat 151:295–307. https://doi.org/10.1007/s10549-015-3395-2
Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, Miyazawa K (2014) Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 289:27386–27399
Yue PJ, Sun YY, Li YH, Xu ZM, Fu WN (2020) MYCT1 inhibits the EMT and migration of laryngeal cancer cells via the SP1/miR-629-3p/ESRP2 pathway. Cell Signal 74:109709. https://doi.org/10.1016/j.cellsig.2020.109709
Mizutani A, Koinuma D, Seimiya H, Miyazono K (2016) The Arkadia-ESRP2 axis suppresses tumor progression: analyses in clear-cell renal cell carcinoma. Oncogene 35:3514–3523. https://doi.org/10.1038/onc.2015.412
Lu ZX, Huang Q, Park JW, Shen S, Lin L, Tokheim CJ, Henry MD, Xing Y (2015) Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol Cancer Res 13:305–318. https://doi.org/10.1158/1541-7786.MCR-14-0366
Fici P, Gallerani G, Morel AP, Mercatali L, Ibrahim T, Scarpi E, Amadori D, Puisieux A, Rigaud M, Fabbri F (2017) Splicing factor ratio as an index of epithelial-mesenchymal transition and tumor aggressiveness in breast cancer. Oncotarget 8:2423–2436. https://doi.org/10.18632/oncotarget.13682
Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, Tolosano E, Provero P, Pandolfi PP, Silengo L, Altruda F (2013) The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS One 8:e72300. https://doi.org/10.1371/journal.pone.0072300
Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, Siziopikou KP, Peng J, Xiao X, Cheng C (2014) Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev 28:1191–1203. https://doi.org/10.1101/gad.241968.114
Sun H, Liu T, Zhu D, Dong X, Liu F, Liang X, Chen C, Shao B, Wang M, Wang Y (2017) HnRNPM and CD44s expression affects tumor aggressiveness and predicts poor prognosis in breast cancer with axillary lymph node metastases. Genes Chromosomes Cancer 56:598–607. https://doi.org/10.1002/gcc.22463
Harvey SE, Xu Y, Lin X, Gao XD, Qiu Y, Ahn J, Xiao X, Cheng C (2018) Coregulation of alternative splicing by hnRNPM and ESRP1 during EMT. RNA 24:1326–1338. https://doi.org/10.1261/rna.066712.118
Hayes GM, Carrigan PE, Miller LJ (2007) Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res 67:2072–2080. https://doi.org/10.1158/0008-5472.CAN-06-2969
Gökmen-Polar Y, Neelamraju Y, Goswami CP, Gu X, Nallamothu G, Janga SC, Badve S (2015) Expression levels of SF3B3 correlate with prognosis and endocrine resistance in estrogen receptor-positive breast cancer. Mod Pathol 28:677–685. https://doi.org/10.1038/modpathol.2014.146
Gökmen-Polar Y, Neelamraju Y, Goswami CP, Gu Y, Gu X, Nallamothu G, Vieth E, Janga SC, Ryan M, Badve SS (2019) Splicing factor ESRP1 controls ER-positive breast cancer by altering metabolic pathways. EMBO Rep 20:e46078. https://doi.org/10.15252/embr.201846078
Vengoji R, Macha MA, Nimmakayala RK, Rachagani S, Siddiqui JA, Mallya K, Gorantla S, Jain M, Ponnusamy MP, Batra SK, Shonka N (2019) Afatinib and Temozolomide combination inhibits tumorigenesis by targeting EGFRvIII-cMet signaling in glioblastoma cells. J Exp Clin Cancer Res 38:266. https://doi.org/10.1186/s13046-019-1264-2
Hagen RM, Ladomery MR (2012) Role of splice variants in the metastatic progression of prostate cancer. Biochem Soc Trans 40:870–874. https://doi.org/10.1042/BST20120026
Seshacharyulu P, Ponnusamy MP, Haridas D, Jain M, Ganti AK, Batra SK (2012) Targeting the EGFR signaling pathway in cancer therapy. Expert Opin Ther Targets 16:15–31. https://doi.org/10.1517/14728222.2011.648617
Urbanski LM, Leclair N, Anczuków O (2018) Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 9:e1476. https://doi.org/10.1002/wrna.1476
Silden E, Hjelle SM, Wergeland L, Sulen A, Andresen V, Bourdon JC, Micklem DR, McCormack E, Gjertsen BT (2013) Expression of TP53 isoforms p53β or p53γ enhances chemosensitivity in TP53null cell lines. PLoS One 8:e56276. https://doi.org/10.1371/journal.pone.0056276
Surget S, Khoury MP, Bourdon JC (2013) Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. Onco Targets Ther 7:57–68. https://doi.org/10.2147/OTT.S53876
Arsic N, Gadéa G, Lagerqvist EL, Busson M, Cahuzac N, Brock C, Hollande F, Gire V, Pannequin J, Roux P (2015) The p53 isoform Δ133p53β promotes cancer stem cell potential. Stem Cell Reports 4:531–540. https://doi.org/10.1016/j.stemcr.2015.02.001
Kourtidis A, Ngok SP, Anastasiadis PZ (2013) p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog Mol Biol Transl Sci 116:409–432. https://doi.org/10.1016/B978-0-12-394311-8.00018-2
Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ (2008) A p120 catenin isoform switch affects rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem 283:18344–18354. https://doi.org/10.1074/jbc.M801192200
Lin JC (2017) Therapeutic applications of targeted alternative splicing to cancer treatment. Int J Mol Sci 19:75. https://doi.org/10.3390/ijms19010075
Orian-Rousseau V, Ponta H (2015) Perspectives of CD44 targeting therapies. Arch Toxicol 89:3–14. https://doi.org/10.1007/s00204-014-1424-2
Li L, Hao X, Qin J, Tang W, He F, Smith A, Zhang M, Simeone DM, Qiao XT, Chen ZN, Lawrence TS, Xu L (2014) Antibody against CD44s inhibits pancreatic tumor initiation and postradiation recurrence in mice. Gastroenterology 146:1108–1118. https://doi.org/10.1053/j.gastro.2013.12.035
Gunia S, Hussein S, Radu DL, Pütz KM, Breyer R, Hecker H, Samii M, Walter GF, Stan AC (1999) CD44s-targeted treatment with monoclonal antibody blocks intracerebral invasion and growth of 9L gliosarcoma. Clin Exp Metastasis 17:221–230. https://doi.org/10.1023/a:1006699203287
Chen J, Lee BH, Williams IR, Kutok JL, Mitsiades CS, Duclos N, Cohen S, Adelsperger J, Okabe R, Coburn A, Moore S, Huntly BJP, Fabbro D, Anderson KC, Griffin JD, Gilliland DG (2005) FGFR3 as a therapeutic target of the small molecule inhibitor PKC412 in hematopoietic malignancies. Oncogene 24:8259–8267. https://doi.org/10.1038/sj.onc.1208989
Bruno IG, Jin W, Cote GJ (2004) Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum Mol Genet 13:2409–2420. https://doi.org/10.1093/hmg/ddh272
Yadav S, Bhagat SD, Gupta A, Samaiya A, Srivastava A, Shukla S (2019) Dietary-phytochemical mediated reversion of cancer-specific splicing inhibits Warburg effect in head and neck cancer. BMC Cancer 19:1031. https://doi.org/10.1186/s12885-019-6257-1
Funding
The authors do not have any funding agency to acknowledge for this review article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Roy Burman, D., Das, S., Das, C. et al. Alternative splicing modulates cancer aggressiveness: role in EMT/metastasis and chemoresistance. Mol Biol Rep 48, 897–914 (2021). https://doi.org/10.1007/s11033-020-06094-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-06094-y