Abstract
Pathogen analysis in wild great apes is both time- and resource-consuming. Therefore, we examined the potential use of urinary neopterin, a sensitive marker of cell-mediated immune system activation, as a disease marker and unspecific screening tool to facilitate informed pathogen analysis in great ape health monitoring. To test this, urinary neopterin was correlated to other disease markers such as sickness behaviors, fever, and urine parameters. Seasonal variation in urinary neopterin levels was investigated as well. The study encompassed noninvasively collected longitudinal data of young wild chimpanzees from the Taï National Park, Côte d´Ivoire. Relationships between disease markers were examined using a linear mixed model and a case study approach. Seasonal variation in urinary neopterin was tested using a linear mixed model. While the linear mixed model found no obvious relationship between urinary neopterin levels and other disease markers, the case study approach revealed a pattern resembling those found in humans. Urinary neopterin levels indicated seasonal immune system activation peaking in times of low ambient temperatures. We suggest the use of urinary neopterin as an unspecific screening tool in great ape health monitoring to identify relevant samples, individuals, and time periods for selective pathogen analysis and zoonotic risk assessment.
Similar content being viewed by others
Introduction
Infectious diseases are a central issue for great ape survival (Leendertz et al. 2006; Gilardi et al. 2015; Leendertz et al. 2017) as increasing contacts between great apes and humans increase the risk of zoonotic disease transmission (Wolfe et al. 2005). Research on infectious diseases in wild great apes has mainly focused on lethal outbreaks with visible clinical symptoms (Hanamura et al. 2008; Köndgen et al. 2008), and on gastrointestinal parasites (Huffman et al. 1997; Gillespie et al. 2010). However, baseline information on mild diseases in great apes barely exists (Lonsdorf et al. 2011; Terio et al. 2011). Mild diseases, defined here as non-lethal diseases, may have negative short- or long-term effects on individual health. The biological relevance of mild diseases regarding disease susceptibility, variation in individual life history, and occurrence of lethal outbreaks is poorly understood. To study non-human primate health, noninvasive techniques have been used including behavioral observation (Alados and Huffman 2000; Macintosh et al. 2011; Ghai et al. 2015), urinalysis (Kaur and Huffman 2004; Leendertz et al. 2010; MacIntosh et al. 2012), body temperature measurement (Jensen et al. 2009), fecal antibody titers (Liu et al. 2008; Neel et al. 2010; Lantz et al. 2016), parasite determination (Huffman et al. 1997; Clough et al. 2010; Masi et al. 2012), and pathogen identification in feces (Köndgen et al. 2010; Schaumburg et al. 2013).
Neopterin (Neo) is a sensitive marker of the cell-mediated immune response and reflects the degree of immune system activation (Fuchs et al. 1988). It is produced by macrophages upon stimulation by interferon gamma deriving from activated T cells (Huber et al. 1984). Levels of Neo in serum and urine show a significant correlation and are stable in body fluids (Fuchs et al. 1994). In human medicine, determining Neo levels has become a standard procedure in evaluating the status of disease (Fuchs et al. 1992), and has recently received attention in non-human primate research (Heistermann and Higham 2015; Higham et al. 2015; Merkley 2017; Müller et al. 2017). Specifically, Neo levels can be reliably and noninvasively measured in chimpanzee and bonobo urine (Behringer et al. 2017). Neo has been shown to correlate with the severity of symptoms (Horak et al. 1998; Zheng et al. 2005; te Witt et al. 2010) as well as with mortality rates in various human infections (Denz et al. 1990; Ruokonen et al. 2002). In summary, Neo levels are a sensitive marker for the detection of a broad spectrum of infectious diseases stimulating the cell-mediated immune system (Mayersbach et al. 1994), and can be used for an unspecific screening for infections caused by known, unidentified, or newly emerging pathogens (Schennach et al. 1997). As pathogen analysis is time- and resource-consuming, determining Neo levels in noninvasively collected urine samples from wild chimpanzees may aid in identifying individuals, and time periods with pronounced immune system challenge for informed pathogen analysis.
In humans and non-human primates, both Neo levels (Fuchs et al. 1992; Winkler et al. 2003; Behringer et al. 2017) and pathogen prevalence (Masi et al. 2012; De Nys et al. 2013; Mapua et al. 2016) have shown to be higher in younger individuals compared to older ones. This suggests a more frequent and intense immune system challenge in young individuals, presumably due to a lack of acquired immunity. Additionally, young individuals may display disease symptoms more obviously compared to older ones, as they may be less exposed to social pressure. In summary, facilitated observation of disease symptoms and increased disease likelihood in young individuals make them appropriate subjects to monitor diseases in wild chimpanzees.
In this study, we examined the use of urinary neopterin (uNeo) levels as a potential marker to monitor mild diseases in wild chimpanzees by correlating them to measures of other disease markers such as sickness behavior, body temperature, and urine parameters. We tested this correlation by using two approaches. First, we incorporated all markers in a statistical model including several individuals followed over several months to explain uNeo level variation. Secondly, we combined all markers in a detailed case report of an individual that displayed prominent sickness behavior. We expected elevated uNeo levels when sickness behavior was observed. Based on the sickness behavior definition by Hart (1988), we used resting times to detect lethargy and fatigue. Play times were used to test for avoidance of voluntary physical activity (Skinner et al. 2009). Fever, which is unspecific to any animal species or disease (Hart 1988), is associated with immune system activation and was expected to coincide with elevated uNeo levels (Brown et al. 1991). Pathologic alterations in urine, indicative for systemic and urinary tract disorders, were assumed to correlate with elevated uNeo levels. Furthermore, we tested whether uNeo levels can be used as an unspecific screening tool for health monitoring by examining seasonal variations in immune system activation.
Methods
Data Collection
The Taï National Park is located in the western part of Côte d´Ivoire and includes tropical rainforest with two wet and dry seasons per year (Boesch and Boesch-Achermann 2000). Samples and data were collected from two habituated chimpanzee groups of the Taï Chimpanzee Project. The study encompassed longitudinal data of 23 chimpanzees (Pan troglodytes verus) aged between 3 and 12 years. Data were collected from two observers between November 2013 and June 2016. Data collection comprised of weekly sampling of urine samples for uNeo determination and urinalysis (Leendertz et al. 2010), body temperature estimation (Jensen et al. 2009), and documentation of behavioral activities on identified individuals (Table 1). For the case report, data of one individual (SOL) were collected on five consecutive days. Urine was collected from leaves and forest floor, and transported on ice in a thermos flask until return to camp. The samples were then transferred into liquid nitrogen, and subsequently shipped to Germany on dry ice. All samples and data were collected noninvasively, following the hygiene guidelines and ethics standards of the Taï Chimpanzee Project (Gilardi et al. 2015). Sample transport was approved by authorities in charge.
Determination of uNeo
To measure the concentration of Neo in chimpanzee urine, we used a commercially available competitive ELISA designed for the quantitative determination of Neo in human urine, serum, and plasma (IBL International GmbH, RE59321, Germany). The Neo ELISA has been previously validated for the determination of uNeo in chimpanzees (Behringer et al. 2017). In total, 537 urine samples were measured in the Endocrinology laboratory of the Max Planck Institute for Evolutionary Anthropology (Leipzig, Germany). To avoid degradation of Neo in urine samples and sample dilutions, exposure to sunlight and repeated freeze–thaw cycles were minimized. The assay was performed according to the manufacturer’s instructions. In brief, urine samples were thawed, vortexed, and centrifuged. Urine samples were diluted with assay buffer provided by the supplier. Subsequently, 20 µl of the diluted samples, standards, and quality controls from the test kit were pipetted in duplicate. 100 µl of enzyme conjugate and 50 µl of Neo antiserum were added. The plate was incubated in the dark on a plate shaker for 90 min at 500 rpm at room temperature. After incubation, the plate was washed four times with 300 µl of washing buffer. Then, 150 µl of tetramethylbenzidine substrate were added, and the plate was incubated for 10 min at room temperature. The reaction was stopped with a stop solution provided by the test kit and the plate subsequently read at 450 nm. Only samples with less than 10% variance in optical density of duplicates were accepted for analysis. The inter-assay variation for all runs (n = 21) constituted of 10.96% for high, and of 11.83% for low quality controls. The intra-assay variation was 6.21% and 9.97% for high and low, respectively. The mean uNeo concentration was corrected for variation in renal excretion using the specific gravity (SG) as described in Miller et al. (2004), which was measured with a digital handheld refractometer (TR35U, TEC Dr. Volker Schmidt GmbH, Germany). Population SG was 1.023. The uNeo concentration is given in nmol/l corr. SG. With respect to possible excessive dilution with rain water, samples with a SG less than 1.002 were excluded from analysis (n = 21).
Behavioral Observation
Data on individual chimpanzee behavior were collected using a digital handheld device (HP iPAQ 2110, Hewlett—Packard). An ethogram was designed using CyberTracker (CyberTracker Conservation, Cape Town, South Africa) to obtain information on main activities (i.e., resting, traveling, and feeding) and social interactions (e.g., grooming and playing). Behavioral terms employed were based on the definitions proposed by Nishida et al. (2010). Behavioral data were collected continuously from dawn-to-dusk on a single individual per day. Individual daily resting and playing times in relation to the entire daily time spent stationary were considered to control for party-dependent traveling behavior. The daily stationary time was obtained by subtracting the daily time spent traveling from the daily observation time. Stationary daily resting and playing times are hereafter called resting and playing, and given in percent.
Body Temperature Measurement
Body temperatures were noninvasively and indirectly measured by recording the temperature decline in fresh feces with a data logger thermometer (K202, Voltcraft) as described in Jensen et al. (2009). In brief, within 3 min after defecation, the two sensors of the thermometer were centrally placed in solid feces and the measurement started. The fecal temperature was continuously recorded in one-second intervals for 6 min. Temperature values were excluded from analysis when inaccurate method application led to calculations outside the range of homoiotherm body temperature, or there were different values for both sensors (n = 40).
Urinalysis
Urinalysis was performed to screen for the presence of leukocytes, protein, erythrocytes, and hemoglobin using Combur9 dipstick tests from Roche Diagnostics (REF 04510046 040, Mannheim, Germany). Fresh urine was applied on the test strip, and results were semiquantitatively determined by visual reading.
Meteorological Data
Meteorological data were recorded daily with a digital thermo-hygrometer. A rain gauge was used to measure daily precipitation. Climatic factors referred to the mean of 14 days prior to urine collection, accounting for the incubation period of many acute infectious diseases.
Statistical Analysis
All statistical analyses were computed in R (Core Team 2017). Calculations of body temperatures were implemented as described in Jensen et al. (2009). Linear mixed models (LMM) (Baayen 2008) were fitted in R using the lmer function of the R package lme4 (Bates et al. 2015).
We tested whether body temperature, resting and playing, the amount of urinary hemoglobin, erythrocytes, leukocytes, and protein (control predictors) explained the variation in uNeo levels (test predictor) using the LMM. We included age at sampling, sex, group, and sampling time as further fixed effects, and individual and observer as random effects. Random slopes were included for all effects except for sex and group within the individual. Except for sex and group, all variables were z-transformed to a mean of zero; for body temperature, z-transformed residuals were used. The uNeo levels were log-transformed. No collinearity was found among the predictors (Field 2005). The sample size for this model consisted in total of 317 observations of 15 individuals (Table 1).
We tested for seasonality and the influence of rainfall, minimum ambient temperature, minimum humidity, and variation in day temperature (control predictors) on uNeo levels (test predictor) using the LMM. To test for seasonality, we included the sine and cosine of Julian date after dividing it by 365.25 and multiplying by 2 × π (Stolwijk et al. 1999). We included age at sampling, sex, group, and sampling time as further fixed effects and individual as random effect. Random slopes were included for all fixed effects except for sex and group within the individual. Except for sex and group, all variables were z-transformed to a mean of zero. Rainfall was additionally square root transformed before z-transformation. The uNeo levels were log-transformed. We found collinearity for minimum ambient temperature with seasonality. Therefore, we compared a reduced model lacking of seasonality to the full model. The sample size for these models was 516 observations of 23 individuals (Table 1).
We assessed model stability of all models by excluding individuals one at a time. This revealed all models to be fairly stable. Correlations among random slopes and intercept were initially included but appeared unidentifiable (Barr et al. 2013; Matuschek et al. 2017). The assumptions of normally and homogenously distributed residuals of random effects were fulfilled in all models.
Results
Relation of Disease Markers
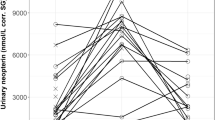
Median uNeo levels varied inter-individually (Fig. 1). Additionally, uNeo levels showed variation within individuals (Figs. 1, 2). The full model compared to the null model did not reveal significance (full: χ2 = 6.338, df = 7, P = 0.501; see Table 2), with uNeo variations in the chimpanzees not obviously explained by any of the test predictors.
Reporting a Mild Disease in a 4-Year-Old Male
We report a case of mild disease in a 4-year-old male chimpanzee (SOL) from January 25 to 29, 2015 (Fig. 2). SOL displayed classic unspecific sickness behavior such as extreme fatigue, lethargy, and inappetence on the 25th. Initially, the time spent resting was remarkably elevated on the 25th, declining over the next few days but still remaining above average resting time (Fig. 3). Play was not observed on 25 and 26 January, but increased toward normal average play rate on the 29th (Fig. 3). On the 25th and 26th, no temperature measurement was performed due to the absence of defecation or diarrhea. A body temperature of 38.4°C was measured on the 27th and 36.9°C and 37.6°C on the following days, respectively. Changes in urine parameters were variably detected for erythrocytes, leukocytes, and hemoglobin from 25 to 28 January (Table 3). In particular, the grade of leukocyturia narrowed down from high to low grade from 25 to 29 January. Sickness behavior was followed by a fourfold increase in uNeo levels, which was measurable 1 day after onset of sickness symptoms, and reached its peak level on 28 January (Fig. 3).
Daily uNeo levels and activity (rest/play) before, during, and after a period of mild disease (25 to 29 January) in a 4-year-old male chimpanzee (SOL). The horizontal line depicts the median uNeo level of SOL. The resting and playing rate of this individual was, on average, 31% and 16% of the day, respectively.
Seasonality of Immune System Activation
The full model was significant when compared to the null model (full: χ2 = 24.852, df = 6, P < 0.0001; see Table 4). Due to collinearity, we then compared a reduced model to the full model, which was also significant (reduced: χ2 = 10.869, df = 2, P = 0.004; Tables 5, 6). Throughout the 3-year study period, we found a seasonal pattern in the uNeo levels with higher levels between November and January compared to other months, reaching highest levels in December and lowest in June (Fig. 4). We found that lower minimum ambient temperatures corresponded with elevated uNeo levels (Table 6).
Discussion
Relation of Disease Markers and Potential Use of uNeo
LMM analysis of sickness behavior, body temperature, and urine parameters did not significantly explain variations in uNeo levels, while the case report found a correlation between uNeo levels and disease markers. Looking at the LMM results based on weekly data collection, uNeo variation was not linked to other disease markers, which may suggest that uNeo is not a suitable or sensitive marker of disease. Additionally, the LMM lacked control of social effects (e.g., availability of play partners and group activities), which may mask sickness behavior. Responsiveness toward disease as well as displaying of sickness behavior within a social context may also vary by individual and was not accounted for in the model. Evidently, the use of uNeo level changes as a noninvasive diagnostic tool is not without limitations. Differences in Neo release have also been observed depending on reinfection rate, endemicity of pathogens (Reibnegger et al. 1987), and pathogen type (Denz et al. 1990). While viral infections cause a remarkable increase in Neo levels (Zheng et al. 2005; Chan et al. 2006), as well as infections with intracellular bacteria (Prat et al. 2006; Cesur et al. 2014), and intracellular parasites (Reibnegger et al. 1984; Brown et al. 1992), Neo levels remain low in extracellular infections (Widner et al. 1999). However, extracellular infections cause changes in disease markers, while Neo levels remain low. This may have limited our model outcome as well. In humans, age is known to impact Neo levels (Fuchs et al. 1992). In our study individuals, age at sampling did not influence uNeo levels. However, the age effect on Neo levels may be more visible when comparing between age classes (e.g., children to adults) rather than within. Plata-Nazar et al. (2007) and Satoh et al. (1998) also found no age dependence when examining Neo levels within children and adolescents. In our study individuals, uNeo levels showed a high degree of intra-individual fluctuation which is found in children as well while levels in adults are stable (Winkler et al. 2003).
When using sickness behavior to define a period of disease, uNeo levels were shown to be elevated as well. uNeo was released with 1 day of delay after onset of disease symptoms, and peaking 3 days after. The kinetics of Neo excretion appears to resemble those in humans (Bloom et al. 1990; Zheng et al. 2005; Chan et al. 2006). A comparable increase in Neo level has been demonstrated when examining the acute immune system response after intravenous endotoxin application in humans (Bloom et al. 1990). Neo rose twofold to fourfold after 24 h of immune system activation and onset of disease symptoms (Bloom et al. 1990). Another study reported a fivefold increase in Neo during acute human SARS infections (Zheng et al. 2005). In our case report, uNeo reached its maximum level 3 days after onset of disease symptoms. Similarly, maximum Neo levels three to 4 days after onset of disease symptoms were reported in humans (Zheng et al. 2005; Chan et al. 2006). According to the human fever definition of Psychrembel (1982), moderate fever of 38.4°C was observed in SOL. We were unable to obtain body temperature measures on all days due to method limitations when diarrhea occurred. Unfortunately, diarrhea often occurs when sick; thus, the fecal measurement is inapplicable. Urinalysis revealed abnormal quantities of hemoglobin, erythrocytes, and leukocytes when uNeo levels were elevated, indicating pathological changes linked to immune system activation. However, urinary dipstick analyses need to be taken with care and may not reflect pathological conditions (Kaur and Huffman 2004; Leendertz et al. 2010; MacIntosh et al. 2012). For instance, the presence of erythrocytes and protein in urine was reported in apparently healthy chimpanzees (Kelly et al. 2004). In our study individuals, elevated levels of urine parameters were also combined with average uNeo levels. In our case study, pathological urine parameters corresponded with elevated uNeo levels. This may be still informative but ultimately requires a larger sample size.
In summary, the case report findings show that sickness behavior, fever, and pathological findings in urine coincide with augmented uNeo levels, suggesting uNeo as a sensitive marker of disease in chimpanzees. While maximum uNeo levels did not correlate with the peak of most pronounced other disease markers, monitoring over several consecutive days revealed a time lag between the onset of disease symptoms and uNeo release. However, given the inter- and intra-individual variation, we emphasize that a longitudinal sampling scheme with weekly data collection per individual may not be suitable to detect and examine correlations between disease markers in chimpanzees. We recommend sampling over consecutive days to detect existing correlations shifted with time.
Seasonality and Unspecific uNeo Screening in Health Monitoring
According to Dowell (2001), seasonality of infectious diseases is formed by several factors ranging across environmental, host, and pathogen-specific factors. Variation in these factors causes complex interactions with others and may lead to changes including transmission or susceptibility of the host. The temporal dispersion of former disease outbreaks in Taï chimpanzees (Köndgen et al. 2008, 2010) might lead to the assumption of seasonally varying underlying mechanisms. We demonstrated seasonal cell-mediated immune activation in young chimpanzees corresponding to uNeo levels with yearly pronounced immune system activation found from November to January. Merkley (2017) reported seasonally determined pattern of cell-mediated immune activation with minimum neopterin levels during the wet season in Kanyawara chimpanzees, which is in line with our results. Environmental factors, which are presumed to foster disease (summarized in Altizer et al. (2006)), may influence the immune system in chimpanzees living in a seasonal environment. When testing them for their potential impact, we found the minimum ambient temperature to have a significant negative effect on uNeo levels. Our results suggest cold stress to be a factor influencing seasonal immune system activation. Cold stress can force energy trade-offs toward thermoregulation, resulting in a compromised immune system (Demas et al. 1997; Svensson et al. 1998; Nelson 2004). Glucocorticoids are released upon stimulation of the HPA axis to adequately respond to changes in metabolic demands, and are known to have immunosuppressive effects (reviewed in Munck et al. (1984)), resulting in increased host susceptibility (Dhabhar 2009). Seasonal trade-offs in immunity attributed to cold stress have been found in several species, leading to variability in susceptibility toward pathogens (Mann et al. 2000; McFarlane et al. 2012; Brown et al. 2016).
Another explanation, which is not mutually exclusive, could be related to group size and sociality. Periods of elevated uNeo levels also coincided with times of food abundance, when Taï chimpanzees are known to form large groups (Kühl et al. 2008). Play rates in infant chimpanzees are increased during months of food abundance compared to other months (Kühl et al. 2008). Group size and social connectivity are positively associated with pathogen transmission (Kühl et al. 2008; Patterson and Ruckstuhl 2013); thus, the social component in infectious disease transmission should not be neglected in the future.
Currently, informed sample selection for pathogen analysis is based on observations of disease symptoms, which are rarely observed in wild chimpanzees (Huffman et al. 1997; Boesch 2008). For instance, one of the study individuals showed extremely elevated uNeo levels 2 days after intense play contact with a moribund conspecific but no sickness behavior was observed. Another individual showed elevated uNeo levels 1 week before its death but displayed no sickness behavior. This rather illustrates the importance of introducing uNeo determination as an unspecific screening tool into great ape health monitoring as sickness behavior may not be reliably observed. Therefore, uNeo seems to be a good candidate to bridge this gap as it covers a broad spectrum of viral, intracellular bacterial, and parasitic pathogens stimulating the cell-mediated immune response. Measuring uNeo levels can particularly help in detecting disease processes with subclinical manifestation such as SIV infection (Fendrich et al. 1989). Noninvasive uNeo screening can aid in identifying relevant samples, individuals, and disease periods for selective pathogen analysis in chimpanzees, and is suitable to study frequency, periods, and patterns of mild diseases and their effects on chimpanzee health on an individual level.
Conclusion
In rural African settings, the increasing contact between great apes and humans due to habitat loss, forest fragmentation, and poaching promotes the transmission of infectious diseases with zoonotic potential (Wolfe et al. 2005). Great apes act as sentinels for emerging infectious diseases (Calvignac-Spencer et al. 2012). Thus, monitoring health changes in great ape populations is therefore essential for both primate and human health. This can provide the basis for effective analysis of disease prevalence, dynamics, and emerging pathogens and assist in making informed decisions regarding future threats toward great ape and human populations.
Finding an appropriate marker to monitor diseases in wild chimpanzees also contributes toward understanding general disease mechanisms in great apes. To account for the temporally dynamic interplay of immune system activation and disease symptoms, a case-oriented study design with consecutive daily sampling is appropriate. Determining uNeo facilitates the comparison of general immune system activation and existing disease dynamics, as well as the identification of risk factors within and across primate populations. We suggest the use of uNeo as an unspecific screening tool in great ape health monitoring to identify relevant samples, individuals, and time periods for selective pathogen analysis and zoonotic risk assessment.
References
Alados, C. L., and M. A. Huffman. 2000. Fractal Long-Range Correlations in Behavioural Sequences of Wild Chimpanzees: a Non-Invasive Analytical Tool for the Evaluation of Health. Ethology 106:105-116.
Altizer, S., A. Dobson, P. Hosseini, P. Hudson, M. Pascual, and P. Rohani. 2006. Seasonality and the dynamics of infectious diseases. Ecology Letters 9:467-484.
Baayen, R. 2008. Analyzing Linguistic Data. Cambridge University Press, University of Alberta.
Barr, D. J., R. Levy, C. Scheepers, and H. J. Tily. 2013. Random effects structure for confirmatory hypothesis testing: Keep it maximal. Journal of memory and language 68 (https://doi.org/10.1016/j.jml.2012.1011.1001)
Bates, D., M. Mächler, B. Bolker, and S. Walker. 2015. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software 67:1-48
Behringer, V., J. M. Stevens, F. H. Leendertz, G. Hohmann, and T. Deschner. 2017. Validation of a Method for the Assessment of Urinary Neopterin Levels to Monitor Health Status in Non-human-primate Species. Frontiers in Physiology 8:51.
Bloom, J. N., A. F. Suffredini, J. E. Parrillo, and A. C. Palestine. 1990. Serum neopterin levels following intravenous endotoxin administration to normal humans. Immunobiology 181:317-323.
Boesch, C. 2008. Why do chimpanzees die in the forest? The challenges of understanding and controlling for wild ape health. American Journal of Primatology 70:722-726.
Boesch, C., and H. Boesch-Achermann. 2000. The Chimpanzees of the Taï Forest: Behavioural Ecology and Evolution. Oxford University Press, Oxford.
Brown, A. E., D. A. Herrington, H. K. Webster, D. F. Clyde, M. B. Sztein, J. R. Davis, M. S. Beier, and R. Edelman. 1992. Urinary neopterin in volunteers experimentally infected with Plasmodium falciparum. Transactions of the Royal Society of Tropical Medicine and Hygiene 86:134-136.
Brown, A. E., P. Teja-Isavadharm, and H. K. Webster. 1991. Macrophage activation in vivax malaria: fever is associated with increased levels of neopterin and interferon-gamma. Parasite Immunology 13:673-679.
Brown, M., P. Hablutzel, I. M. Friberg, A. G. Thomason, A. Stewart, J. A. Pachebat, and J. A. Jackson. 2016. Seasonal immunoregulation in a naturally-occurring vertebrate. BMC Genomics 17:369.
Calvignac-Spencer, S., S. A. Leendertz, T. R. Gillespie, and F. H. Leendertz. 2012. Wild great apes as sentinels and sources of infectious disease. Clinical Microbiology and Infection 18:521-527.
Cesur, S., T. Aslan, N. T. Hoca, F. Cimen, G. Tarhan, A. Cifci, I. Ceyhan, and T. Sipit. 2014. Clinical importance of serum neopterin level in patients with pulmonary tuberculosis. Int J Mycobacteriol 3:5-8.
Chan, C. P., J. W. Choi, K. Y. Cao, M. Wang, Y. Gao, D. H. Zhou, B. Di, H. F. Xu, M. F. Leung, A. Bergmann, M. Lehmann, Y. M. Nie, G. W. Cautherley, D. Fuchs, R. Renneberg, and B. J. Zheng. 2006. Detection of serum neopterin for early assessment of dengue virus infection. Journal of Infection 53:152-158.
Clough, D., M. Heistermann, and P. M. Kappeler. 2010. Host intrinsic determinants and potential consequences of parasite infection in free-ranging red-fronted lemurs (Eulemur fulvus rufus). American Journal of Physical Anthropology 142:441-452.
De Nys, H. M., S. Calvignac-Spencer, U. Thiesen, C. Boesch, R. M. Wittig, R. Mundry, and F. H. Leendertz. 2013. Age-related effects on malaria parasite infection in wild chimpanzees. Biology Letters 9:20121160.
Demas, G. E., V. Chefer, M. I. Talan, and R. J. Nelson. 1997. Metabolic costs of mounting an antigen-stimulated immune response in adult and aged C57BL/6 J mice. American Journal of Physiology 273:R1631-1637.
Denz, H., D. Fuchs, A. Hausen, H. Huber, D. Nachbaur, G. Reibnegger, J. Thaler, E. R. Werner, and H. Wachter. 1990. Value of urinary neopterin in the differential diagnosis of bacterial and viral infections. Klinische Wochenschrift 68:218-222.
Dhabhar, F. S. 2009. Enhancing versus Suppressive Effects of Stress on Immune Function: Implications for Immunoprotection and Immunopathology. Neuroimmunomodulation 16:300-317.
Dowell, S. F. 2001. Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerging Infectious Diseases 7:369-374.
Fendrich, C., W. Luke, C. Stahl-Hennig, O. Herchenroder, D. Fuchs, H. Wachter, and G. Hunsmann. 1989. Urinary neopterin concentrations in rhesus monkeys after infection with simian immunodeficiency virus (SIVmac 251). AIDS 3:305-307.
Field, A. 2005. Discovering Statistics Using SPSS. Sage Publications Ltd., London.
Fuchs, D., A. Hausen, G. Reibnegger, E. R. Werner, M. P. Dierich, and H. Wachter. 1988. Neopterin as a marker for activated cell-mediated immunity: application in HIV infection. Immunology Today 9:150-155.
Fuchs, D., C. Stahl-Hennig, A. Gruber, C. Murr, G. Hunsmann, and H. Wachter. 1994. Neopterin–its clinical use in urinalysis. Kidney International 47:S8-11.
Fuchs, D., G. Weiss, G. Reibnegger, and H. Wachter. 1992. The role of neopterin as a monitor of cellular immune activation in transplantation, inflammatory, infectious, and malignant diseases. Critical Reviews in Clinical Laboratory Sciences 29:307-341.
Ghai, R. R., V. Fugere, C. A. Chapman, T. L. Goldberg, and T. J. Davies. 2015. Sickness behaviour associated with non-lethal infections in wild primates. Proceedings Biological Sciences 282:20151436.
Gilardi, K. V., T. R. Gillespie, F. H. Leendertz, E. J. Macfie, D. A. Travis, C. A. Whittier, and E. A. Williamson. 2015. Best Practice Guidelines for Health Monitoring and Disease Control in Great Ape Populations. Gland, Switzerland: IUCN SSC Primate Specialist Group
Gillespie, T. R., E. V. Lonsdorf, E. P. Canfield, D. J. Meyer, Y. Nadler, J. Raphael, A. E. Pusey, J. Pond, J. Pauley, T. Mlengeya, and D. A. Travis. 2010. Demographic and ecological effects on patterns of parasitism in eastern chimpanzees (Pan troglodytes schweinfurthii) in Gombe National Park, Tanzania. American Journal of Physical Anthropology 143:534-544
Hanamura, S., M. Kiyono, M. Lukasik-Braum, T. Mlengeya, M. Fujimoto, M. Nakamura, and T. Nishida. 2008. Chimpanzee deaths at Mahale caused by a flu-like disease. Primates 49:77-80.
Hart, B. L. 1988. Biological basis of the behavior of sick animals. Neuroscience & Biobehaviral Reviews 12:123-137.
Heistermann, M., and J. P. Higham. 2015. Urinary neopterin, a non-invasive marker of mammalian cellular immune activation, is highly stable under field conditions. Scientific Reports 5:16308.
Higham, J. P., C. Kraus, C. Stahl-Hennig, A. Engelhardt, D. Fuchs, and M. Heistermann. 2015. Evaluating noninvasive markers of nonhuman primate immune activation and inflammation. American Journal of Physical Anthropology 158:673-684.
Horak, E., I. Gassner, B. Solder, H. Wachter, and D. Fuchs. 1998. Neopterin levels and pulmonary tuberculosis in infants. Lung 176:337-344.
Huber, C., J. R. Batchelor, D. Fuchs, A. Hausen, A. Lang, D. Niederwieser, G. Reibnegger, P. Swetly, J. Troppmair, and H. Wachter. 1984. Immune response-associated production of neopterin. Release from macrophages primarily under control of interferon-gamma. Journal of Experimental Medicine 160:310-316.
Huffman, M. A., S. Gotoh, L. A. Turner, M. Hamai, and K. Yoshida. 1997. Seasonal trends in intestinal nematode infection and medicinal plant use among chimpanzees in the Mahale Mountains, Tanzania. Primates 38:111-125.
Jensen, S. A., R. Mundry, C. L. Nunn, C. Boesch, and F. H. Leendertz. 2009. Non-invasive body temperature measurement of wild chimpanzees using fecal temperature decline. Journal of Wildlife Diseases 45:542-546.
Kaur, T., and M. A. Huffman. 2004. Descriptive urological record of chimpanzees (Pan troglodytes) in the wild and limitations associated with using multi-reagent dipstick test strips. Journal of Medical Primatology 33:187-196.
Kelly, T. R., J. M. Sleeman, and R. Wrangham. 2004. Urinalysis in free-living chimpanzees (Pan troglodytes schweinfurthii) in Uganda. Veterinary Record 154:729-730.
Köndgen, S., H. Kühl, P. K. N’Goran, P. D. Walsh, S. Schenk, N. Ernst, R. Biek, P. Formenty, K. Matz-Rensing, B. Schweiger, S. Junglen, H. Ellerbrok, A. Nitsche, T. Briese, W. I. Lipkin, G. Pauli, C. Boesch, and F. H. Leendertz. 2008. Pandemic human viruses cause decline of endangered great apes. Current Biology 18:260-264.
Köndgen, S., S. Schenk, G. Pauli, C. Boesch, and F. H. Leendertz. 2010. Noninvasive monitoring of respiratory viruses in wild chimpanzees. Ecohealth 7:332-341.
Kühl, H. S., C. Elzner, Y. Moebius, C. Boesch, and P. D. Walsh. 2008. The price of play: self-organized infant mortality cycles in chimpanzees. PLoS One 3:e2440.
Lantz, E. L., E. V. Lonsdorf, M. R. Heintz, C. M. Murray, I. Lipende, D. A. Travis, and R. M. Santymire. 2016. Non-invasive quantification of immunoglobulin A in chimpanzees (Pan troglodytes schweinfurthii) at Gombe National Park, Tanzania. American Journal of Primatology 80:e22558.
Leendertz, F. H., G. Pauli, K. Maetz-Rensing, W. Boardman, C. Nunn, H. Ellerbrok, S. A. Jensen, S. Junglen, and B. Christophe. 2006. Pathogens as drivers of population declines: The importance of systematic monitoring in great apes and other threatened mammals. Biological Conservation 131:325-337.
Leendertz, S. A., S. Metzger, E. Skjerve, T. Deschner, C. Boesch, J. Riedel, and F. H. Leendertz. 2010. A longitudinal study of urinary dipstick parameters in wild chimpanzees (Pan troglodytes verus) in Cote d’Ivoire. American Journal of Primatology 72:689-698.
Leendertz, S. A. J., S. A. Wich, M. Ancrenaz, R. A. Bergl, M. K. Gonder, T. Humle, and F. H. Leendertz. 2017. Ebola in great apes – current knowledge, possibilities for vaccination, and implications for conservation and human health. Mammal Review 47:98-111.
Liu, W., M. Worobey, Y. Li, B. F. Keele, F. Bibollet-Ruche, Y. Guo, P. A. Goepfert, M. L. Santiago, J.-B. N. Ndjango, C. Neel, S. L. Clifford, C. Sanz, S. Kamenya, M. L. Wilson, A. E. Pusey, N. Gross-Camp, C. Boesch, V. Smith, K. Zamma, M. A. Huffman, J. C. Mitani, D. P. Watts, M. Peeters, G. M. Shaw, W. M. Switzer, P. M. Sharp, and B. H. Hahn. 2008. Molecular Ecology and Natural History of Simian Foamy Virus Infection in Wild-Living Chimpanzees. PLoS Pathogens 4:e1000097.
Lonsdorf, E. V., C. M. Murray, E. V. Lonsdorf, D. A. Travis, I. C. Gilby, J. Chosy, J. Goodall, and A. E. Pusey. 2011. A retrospective analysis of factors correlated to chimpanzee (Pan troglodytes schweinfurthii) respiratory health at Gombe National Park, Tanzania. Ecohealth 8:26-35.
Macintosh, A. J., C. L. Alados, and M. A. Huffman. 2011. Fractal analysis of behaviour in a wild primate: behavioural complexity in health and disease. Journal of the Royal Society Interface 8:1497-1509.
MacIntosh, A. J. J., M. A. Huffman, K. Nishiwaki, and T. Miyabe-Nishiwaki. 2012. Urological Screening of a Wild Group of Japanese Macaques (Macaca fuscata yakui): Investigating Trends in Nutrition and Health. International Journal of Primatology 33:460-478.
Mann, D. R., M. A. Akinbami, K. G. Gould, and A. A. Ansari. 2000. Seasonal variations in cytokine expression and cell-mediated immunity in male rhesus monkeys. Cellular Immunology 200:105-115.
Mapua, M. I., K. J. Petrzelkova, J. Burgunder, E. Dadakova, K. Brozova, K. Hrazdilova, F. A. Stewart, A. K. Piel, P. Vallo, H. P. Fuehrer, C. Hashimoto, D. Modry, and M. A. Qablan. 2016. A comparative molecular survey of malaria prevalence among Eastern chimpanzee populations in Issa Valley (Tanzania) and Kalinzu (Uganda). Malaria Journal 15:423.
Masi, S., S. Chauffour, O. Bain, A. Todd, J. Guillot, and S. Krief. 2012. Seasonal effects on great ape health: a case study of wild chimpanzees and Western gorillas. PLoS One 7:e49805.
Matuschek, H., R. Kliegl, S. Vasishth, H. Baayen, and D. Bates. 2017. Balancing Type I Error and Power in Linear Mixed Models. Journal of memory and language 94:305–315.
Mayersbach, P., R. Augustin, H. Schennach, D. Schonitzer, E. R. Werner, H. Wachter, and G. Reibnegger. 1994. Commercial enzyme-linked immunosorbent assay for neopterin detection in blood donations compared with RIA and HPLC. Clinical Chemistry 40:265-266.
McFarlane, D., R. F. Wolf, K. A. McDaniel, and G. L. White. 2012. The effect of season on inflammatory response in captive baboons. Journal of Medical Primatology 41:341-348.
Merkley S (2017) Ecological Immunology: Neopterin as a Non-invasive Biomarker for Longitudinal Analysis of Immune Life Histories in Primates. Master. University of New Mexico, unpublished
Miller, R. C., E. Brindle, D. J. Holman, J. Shofer, N. A. Klein, M. R. Soules, and K. A. O’Connor. 2004. Comparison of specific gravity and creatinine for normalizing urinary reproductive hormone concentrations. Clinical Chemistry 50:924-932.
Müller, N., M. Heistermann, C. Strube, O. Schülke, and J. Ostner. 2017. Age, but not anthelmintic treatment, is associated with urinary neopterin levels in semi-free ranging Barbary macaques. Scientific Reports 7:41973.
Munck, A., P. M. Guyre, and N. J. Holbrook. 1984. Physiological functions of glucocorticoids in stress and their relation to pharmacological actions. Endocrine Reviews 5:25-44.
Neel, C., L. Etienne, Y. Li, J. Takehisa, R. S. Rudicell, I. N. Bass, J. Moudindo, A. Mebenga, A. Esteban, F. Van Heuverswyn, F. Liegeois, P. J. Kranzusch, P. D. Walsh, C. M. Sanz, D. B. Morgan, J. B. Ndjango, J. C. Plantier, S. Locatelli, M. K. Gonder, F. H. Leendertz, C. Boesch, A. Todd, E. Delaporte, E. Mpoudi-Ngole, B. H. Hahn, and M. Peeters. 2010. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. Journal of Virology 84:1464-1476.
Nelson, R. J. 2004. Seasonal immune function and sickness responses. Trends in Immunology 25:187-192.
Nishida, T., K. Zamma, T. Matsusaka, A. Inaba, and W. C. McGrew. 2010. Chimpanzee Behavior in The Wild. Page 227 in T. Nishida, K. Zamma, T. Matsusaka, A. Inaba, and W. C. McGrew, editors. Chimpanzee Behavior in The Wild. Springer, Tokyo
Patterson, J. E., and K. E. Ruckstuhl. 2013. Parasite infection and host group size: a meta-analytical review. Parasitology 140:803-813.
Plata-Nazar, K., G. Luczak, A. Borkowska, A. Delinska-Galinska, E. Kozielska, K. Marek, and M. Korzon. 2007. Reference Standard of Serum Neopterin Concentration in Healthy Children. Page 19 Pteridines
Prat, C., J. Dominguez, F. Andreo, S. Blanco, A. Pallares, F. Cuchillo, C. Ramil, J. Ruiz-Manzano, and V. Ausina. 2006. Procalcitonin and neopterin correlation with aetiology and severity of pneumonia. Journal of Infection 52:169-177.
Psychrembel, W. 1982. Psychrembel Klinisches Wörterbuch. Page 1341 in W. Psychrembel, editor. Psychrembel Klinisches Wörterbuch. Walter de Gruyter, Berlin.
R Core Team (2017) Version 3.3.3, R Core Team (2017), R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/
Reibnegger, G., V. Boonpucknavig, D. Fuchs, A. Hausen, E. Schmutzhard, and H. Wachter. 1984. Urinary neopterin is elevated in patients with malaria. Transactions of the Royal Society of Tropical Medicine and Hygiene 78:545-546.
Reibnegger, G., D. Fuchs, A. Hausen, E. Schmutzhard, E. R. Werner, and H. Wachter. 1987. The dependence of cell-mediated immune activation in malaria on age and endemicity. Transactions of the Royal Society of Tropical Medicine and Hygiene 81:729-733.
Ruokonen, E., L. Ilkka, M. Niskanen, and J. Takala. 2002. Procalcitonin and neopterin as indicators of infection in critically ill patients. Acta Anaesthesiologica Scandinavica 46:398-404.
Satoh, T., L. M. Brown, W. A. Blattner, E. M. Maloney, C. C. Kurman, D. L. Nelson, D. Fuchs, H. Wachter, and D. J. Tollerud. 1998. Serum neopterin, beta2-microglobulin, soluble interleukin-2 receptors, and immunoglobulin levels in healthy adolescents. Clinical Immunology and Immunopathology 88:176-182.
Schaumburg, F., L. Mugisha, P. Kappeller, C. Fichtel, R. Kock, S. Kondgen, K. Becker, C. Boesch, G. Peters, and F. Leendertz. 2013. Evaluation of non-invasive biological samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS One 8:e78046.
Schennach, H., D. Schonitzer, H. Wachter, and D. Fuchs. 1997. Blood donations and viruses. The Lancet 349:1327-1328.
Skinner, G. W., D. Mitchell, and L. M. Harden. 2009. Avoidance of physical activity is a sensitive indicator of illness. Physiology and Behavior 96:421-427.
Stolwijk, A. M., H. Straatman, and G. A. Zielhuis. 1999. Studying seasonality by using sine and cosine functions in regression analysis. Journal of Epidemiology and Community Health 53:235-238.
Svensson, E., L. RÅberg, C. Koch, and D. Hasselquist. 1998. Energetic stress, immunosuppression and the costs of an antibody response. Functional Ecology 12:912-919.
te Witt, R., M. E. van Wolfswinkel, P. L. Petit, J. J. van Hellemond, R. Koelewijn, A. van Belkum, and P. J. van Genderen. 2010. Neopterin and procalcitonin are suitable biomarkers for exclusion of severe Plasmodium falciparum disease at the initial clinical assessment of travellers with imported malaria. Malaria Journal 9:255.
Terio, K. A., M. J. Kinsel, J. Raphael, T. Mlengeya, I. Lipende, C. A. Kirchhoff, B. Gilagiza, M. L. Wilson, S. Kamenya, J. D. Estes, B. F. Keele, R. S. Rudicell, W. Liu, S. Patton, A. Collins, B. H. Hahn, D. A. Travis, and E. V. Lonsdorf. 2011. Pathologic lesions in chimpanzees (Pan trogylodytes schweinfurthii) from Gombe National Park, Tanzania, 2004-2010. Journal of Zoo and Wildlife Medicine 42:597-607.
Widner, B., C. Murr, B. Wirleitner, C. Mayr, N. Spöttl, G. Baier-Bitterlich, and D. Fuchs. 1999. The Importance of Neopterin as a Laboratory Diagnostic Marker of Immune Activation. Page 101 Pteridines
Winkler, C., B. Wirleitner, E. R. Werner, and D. Fuchs. 2003. Urinary neopterin concentrations in healthy individuals with household contact. Pteridines 14:34-38.
Wolfe, N. D., P. Daszak, A. M. Kilpatrick, and D. S. Burke. 2005. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerging Infectious Diseases 11:1822-1827.
Zheng, B., K. Y. Cao, C. P. Chan, J. W. Choi, W. Leung, M. Leung, Z. H. Duan, Y. Gao, M. Wang, B. Di, J. M. Hollidt, A. Bergmann, M. Lehmann, I. Renneberg, J. S. Tam, P. K. Chan, G. W. Cautherley, D. Fuchs, and R. Renneberg. 2005. Serum neopterin for early assessment of severity of severe acute respiratory syndrome. Clinical Immunology 116:18-26.
Acknowledgements
We thank the Ministry of Environment, Water Resources and Forests, the Ministry of Research, the Office Ivoirien des Parcs et Réserves, and the Centre Suisse de Recherches Scientifiques in Côte d´Ivoire for research permission and logistical support. We thank the staff of the Taï Chimpanzee Project, especially the field assistant Oulai Landry for help in the field. We thank Roger Mundry for statistical support, and Ines Hirschberg and Anna Kraft for technical support. This research was funded by the Max Planck Society, the German Academic Exchange Service, and the Deutsche Forschungsgemeinschaft, the framework of the research group “Sociality and Health in Primates” (DFG FOR 2136).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Löhrich, T., Behringer, V., Wittig, R.M. et al. The Use of Neopterin as a Noninvasive Marker in Monitoring Diseases in Wild Chimpanzees. EcoHealth 15, 792–803 (2018). https://doi.org/10.1007/s10393-018-1357-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10393-018-1357-y