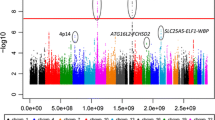
Abstract
Background
There are substantial differences in inflammatory bowel disease (IBD) genetics depending on the populations examined. We aimed to identify Japanese population-specific or true culprit susceptibility genes through a meta-analysis of past genetic studies of Japanese IBD.
Methods
For this study, we reviewed 2,703 articles. The review process consisted of three screening stages: we initially searched for relevant studies and then relevant single nucleotide polymorphisms (SNPs). Finally, we adjusted them for the meta-analysis. To maximize our chances of analysis, we introduced proxy SNPs during the first stage. To minimize publication bias, no significant SNPs and solitary SNPs without pairs were combined to be reconsidered during the third stage. Additionally, two SNPs were newly genotyped. Finally, we conducted a meta-analysis of 37 published studies in 50 SNPs located at 22 loci corresponding to the total number of 4,853 Crohn’s disease (CD), 5,612 ulcerative colitis (UC) patients, and 14,239 healthy controls.
Results
We confirmed that the NKX2–3 polymorphism is associated with common susceptibility to IBD and that HLA-DRB1*0450 alleles increase susceptibility to CD but reduce risk for UC while HLA-DRB1*1502 alleles increase susceptibility to UC but reduce CD risk. Moreover, we found individual disease risk loci: TNFSF15 and TNFα to CD and HLA-B*5201, and NFKBIL1 to UC. The genetic risk of HLA was substantially high (odds ratios ranged from 1.54 to 2.69) while that of common susceptibility loci to IBD was modest (odds ratio ranged from 1.13 to 1.24).
Conclusions
Results indicate that Japanese IBD susceptibility loci identified by the meta-analysis are closely associated with the HLA regions.






Similar content being viewed by others
References
Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621.
Hibi T, Ogata H. Novel pathophysiological concepts of inflammatory bowel disease. J Gastroenterol. 2006;41:10–6.
Kuwahara E, Asakura K, Nishiwaki Y, Inoue N, Watanabe M, Hibi T, et al. Effect of family history on inflammatory bowel disease characteristics in Japanese patients. J Gastroenterol. 2012;47:961–8.
Van Limbergen J, Russell RK, Nimmo ER, Satsangi J. The genetics of inflammatory bowel disease. Am J Gastroenterol. 2007;102:2820–31.
Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2.
Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–20.
Franke A, Balschun T, Sina C, Ellinghaus D, Häsler R, Mayr G, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL). Nat Genet. 2010;42:292–4.
Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24.
Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603.
Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11.
Steinman L. A brief history of Th17, the first major revision in the Th1/Th2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–45.
Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–8.
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL-23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3.
Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology. 2002;123:86–91.
Yamazaki K, Takazoe M, Tanaka T, Ichimori T, Saito S, Iida A, et al. Association analysis of SLC22A4, SLC22A5 and DLG5 in Japanese patients with Crohn disease. J Hum Genet. 2004;49:664–8.
Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet. 2007;52:575–83.
Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–506.
Asano K, Matsushita T, Umeno J, Hosono N, Takahashi A, Kawaguchi T, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. 2009;41:1325–9.
Okada Y, Yamazaki K, Umeno J, Takahashi A, Kumasaka N, Ashikawa K, et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn’s disease. Gastroenterology. 2011;141:864–71.
Yamazaki K, Umeno J, Takahashi A, Hirano A, Johnson TA, Kumasaka N, et al. A genome-wide association study identifies 2 susceptibility loci for Crohn’s disease in a Japanese population. Gastroenterology. 2013;144:781–8.
Moher D, Liberati A, Tetzlaff J, Altman DG, the PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Wang Z, Hu J, Fan R, Zhou J, Zhong J. Association between CD14 gene C-260T polymorphism and inflammatory bowel disease: a meta-analysis. PLoS ONE. 2012;7:e45144.
Seiderer J, Elben I, Diegelmann J, Glas J, Stallhofer J, Tillack C, et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm Bowel Dis. 2008;14:437–45.
Tanaka M, Arimura Y, Goto A, Hosokawa M, Nagaishi K, Yamashita K, et al. Genetic variants in surfactant, pulmonary-associated protein D (SFTPD) and Japanese susceptibility to ulcerative colitis. Inflamm Bowel Dis. 2009;15:918–25.
Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2–6.
Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291–306.
Breslow NE, Day NE. Statistical methods in cancer research. Volume 1—the analysis of case–control studies. Lyon, UK International Agency for Research on Cancer. IARC Sci Publ. 1980;32:5–338.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Weir BS. Genetic data analysis II. Sunderland: Sinauer Associates; 1996.
Arai T, Kakuta Y, Kinouchi Y, Kimura T, Negoro K, Aihara H, et al. Increased expression of NKX2.3 mRNA transcribed from the risk haplotype for ulcerative colitis in the involved colonic mucosa. Hum Immunol. 2011;72:587–91.
Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, et al. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn’s disease in Japanese patients. Gut. 2009;58:228–32.
Nakagome S, Takeyama Y, Mano S, Sakisaka S, Matsui T, Kawamura S, et al. Population-specific susceptibility to Crohn’s disease and ulcerative colitis; dominant and recessive relative risks in the Japanese population. Ann Hum Genet. 2010;74:126–36.
Kakuta Y, Kinouchi Y, Negoro K, Takahashi S, Shimosegawa T. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut. 2006;55:1527–8.
Prescott NJ, Dominy KM, Kubo M, Lewis CM, Fisher SA, Redon R, et al. Independent and population-specific association of risk variants at the IRGM locus with Crohn’s disease. Hum Mol Genet. 2010;19:1828–39.
Hayashi R, Tahara T, Shiroeda H, Saito T, Nakamura M, Tsutsumi M, et al. Influence of IL17A polymorphisms (rs2275913 and rs3748067) on the susceptibility to ulcerative colitis. Clin Exp Med. 2012;. doi:10.1007/s10238-012-0206-5.
Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28:44–9.
Takagawa T, Tamura K, Takeda N, Tomita T, Ohda Y, Fukunaga K, et al. Association between IL-18 gene promoter polymorphisms and inflammatory bowel disease in a Japanese population. Inflamm Bowel Dis. 2005;11:1038–43.
Tamura K, Fukuda Y, Sashio H, Takeda N, Bamba H, Kosaka T, et al. IL18 polymorphism is associated with an increased risk of Crohn’s disease. J Gastroenterol. 2002;37:111–6.
Sashio H, Tamura K, Ito R, Yamamoto Y, Bamba H, Kosaka T, et al. Polymorphisms of the TNF gene and the TNF receptor superfamily member 1B gene are associated with susceptibility to ulcerative colitis and Crohn’s disease, respectively. Immunogenetics. 2002;53:1020–7.
Negoro K, Kinouchi Y, Hiwatashi N, Takahashi S, Takagi S, Satoh J, et al. Crohn’s disease is associated with novel polymorphisms in the 5′-flanking region of the tumor necrosis factor gene. Gastroenterology. 1999;117:1062–8.
Matsuoka M, Sudo S, Nakamura M, Sugisaka H, Negishi M, Miyakawa Y, et al. Genetic association analysis of TNF-α and CD14 gene polymorphism with inflammatory bowel disease. Jpn J Gastroenterol. 2003;100(Suppl):A711 (in Japanese).
Aoyagi Y, Nagata S, Kudo T, Fujii T, Wada M, Chiba Y, et al. Peroxisome proliferator-activated receptor γ 2 mutation may cause a subset of ulcerative colitis. Pediatr Int. 2010;52:729–34.
Wang F, Tahara T, Arisawa T, Sakata M, Takahama K, Watanabe M, et al. Polymorphism of peroxisome proliferator-activated receptor gamma is not associated to Japanese ulcerative colitis. Hepatogastroenterology. 2008;55:73–5.
Sato K, Shiota M, Fukuda S, Iwamoto E, Machida H, Inamine T, et al. Strong evidence of a combination polymorphism of the tyrosine kinase 2 gene and the signal transducer and activator of transcription 3 gene as a DNA-based biomarker for susceptibility to Crohn’s disease in the Japanese population. J Clin Immunol. 2009;29:815–25.
Machida H, Tsukamoto K, Wen CY, Narumi Y, Shikuwa S, Isomoto H, et al. Association of polymorphic alleles of CTLA4 with inflammatory bowel disease in the Japanese. World J Gastroenterol. 2005;11:4188–93.
Matsuzawa J, Sugimura K, Matsuda Y, Takazoe M, Ishizuka K, Mochizuki T, et al. Association between K469E allele of intercellular adhesion molecule 1 gene and inflammatory bowel disease in a Japanese population. Gut. 2003;52:75–8.
Narumi Y, Isomoto H, Shiota M, Sato K, Kondo S, Machida H, et al. Polymorphisms of PTPN11 coding SHP-2 as biomarkers for ulcerative colitis susceptibility in the Japanese population. J Clin Immunol. 2009;29:303–10.
Osuga T, Sakaeda T, Nakamura T, Yamada T, Koyama T, Tamura T, et al. MDR1 C3435T polymorphism is predictive of later onset of ulcerative colitis in Japanese. Biol Pharm Bull. 2006;29:324–9.
Kosaka T, Yoshino J, Inui K, Wakabayashi T, Okushima K, Kobayashi T, et al. Association between SHMT gene polymorphisms and ulcerative colitis. Jpn J Gastroenterol. 2009;106(Suppl):A826 (in Japanese).
Shiroeda H, Tahara T, Nakamura M, Shibata T, Nomura T, Yamada H, et al. Association between functional promoter polymorphisms of macrophage migration inhibitory factor (MIF) gene and ulcerative colitis in Japan. Cytokine. 2010;51:173–7.
Nohara H, Okayama N, Inoue N, Koike Y, Fujimura K, Suehiro Y, et al. Association of the −173 G/C polymorphism of the macrophage migration inhibitory factor gene with ulcerative colitis. J Gastroenterol. 2004;39:242–6.
Fuse K, Katakura K, Sakamoto N, Ohira H. Toll-like receptor 9 gene mutations and polymorphisms in Japanese ulcerative colitis patients. World J Gastroenterol. 2010;16:5815–21.
Wang F, Tahara T, Arisawa T, Shibata T, Nakamura M, Fujita H, et al. Genetic polymorphisms of CD14 and Toll-like receptor-2 (TLR2) in patients with ulcerative colitis. J Gastroenterol Hepatol. 2007;22:925–9.
Obana N, Takahashi S, Kinouchi Y, Negoro K, Takagi S, Hiwatashi N, et al. Ulcerative colitis is associated with a promoter polymorphism of lipopolysaccharide receptor gene, CD14. Scand J Gastroenterol. 2002;37:699–704.
Oomori S. Association between polymorphisms in the inhibitor of Kappa B-like protein (IKBL) gene and ulcerative colitis in a Japanese population. Med J Sendai Red Cross Hosp. 2009;18:23–32 (in Japanese).
Aizawa H, Kinouchi Y, Negoro K, Nomura E, Imai G, Takahashi S, et al. HLA-B is the best candidate of susceptibility genes in HLA for Japanese ulcerative colitis. Tissue Antigens. 2009;73:569–74.
Kinouchi Y, Matsumoto K, Negoro K, Takagi S, Takahashi S, Hiwatashi N, et al. HLA-B genotype in Japanese patients with Crohn’s disease. Dis Colon Rectum. 2003;46:S10–4.
Seki SS, Sugimura K, Ota M, Matsuzawa J, Katsuyama Y, Ishizuka K, et al. Stratification analysis of MICA triplet repeat polymorphisms and HLA antigens associated with ulcerative colitis in Japanese. Tissue Antigens. 2001;58:71–6.
Futami S, Aoyama N, Honsako Y, Tamura T, Morimoto S, Nakashima T, et al. HLA-DRB1*1502 allele, subtype of DR15, is associated with susceptibility to ulcerative colitis and its progression. Dig Dis Sci. 1995;40:814–8.
Matsumura Y, Kinouchi Y, Nomura E, Negoro K, Kakuta Y, Endo K, et al. HLA-DRB1 alleles influence clinical phenotypes in Japanese patients with ulcerative colitis. Tissue Antigens. 2008;71:447–52.
Tremelling M, Berzuini C, Massey D, Bredin F, Price C, Dawson C, et al. Contribution of TNFSF15 gene variants to Crohn’s disease susceptibility confirmed in UK population. Inflamm Bowel Dis. 2008;14:733–7.
Takedatsu H, Michelsen KS, Wei B, Landers CJ, Thomas LS, Dhall D, et al. TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 2008;135:552–67.
Lawrence PA, Morata G. Homeobox genes: their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–9.
Buchberger A, Pabst O, Brand T, Seidl K, Arnold HH. Chick NKx-2.3 represents a novel family member of vertebrate homologues to the Drosophila homeobox gene tinman: differential expression of cNKx-2.3 and cNKx-2.5 during heart and gut development. Mech Dev. 1996;56:151–63.
Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNk2–3 and XNk2–5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–49.
Wang CC, Biben C, Robb L, Nassir F, Barnett L, Davidson NO, et al. Homeodomain factor Nk2–3 controls regional expression of leukocyte homing coreceptor MAdCAM-1 in specialized endothelial cells of the viscera. Dev Biol. 2000;224:152–67.
Pabst O, Förster R, Lipp M, Engel H, Arnold HH. NKX2.3 is required for MAdCAM-1 expression and homing of lymphocytes in spleen and mucosa-associated lymphoid tissue. EMBO J. 2000;19:2015–23.
Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395–401.
Ahmad T, Marshall SE, Jewell D. Genetics of inflammatory bowel disease: the role of the HLA complex. World J Gastroenterol. 2006;12:3628–35.
Yang SK, Hong M, Zhao W, Jung Y, Baek J, Tayebi N, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2013;. doi:10.1136/gutjnl-2013-305193.
Ng SC, Tsoi KK, Kamm MA, Xia B, Wu J, Chan FK, et al. Genetics of inflammatory bowel disease in Asia: systematic review and meta-analysis. Inflamm Bowel Dis. 2012;6:1164–76.
Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–5.
International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). Ulcerative colitis meta-analysis summary statistics. http://www.ibdgenetics.org/downloads.html.
Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024.
Ahmad T, Armuzzi A, Neville M, Bunce M, Ling KL, et al. The contribution of human leucocyte antigen complex genes to disease phenotype in ulcerative colitis. Tissue Antigens. 2003;62:527–35.
Roussomoustakaki M, Satsangi J, Welsh K, Louis E, Fanning G, et al. Genetic markers may predict disease behavior in patients with ulcerative colitis. Gastroenterology. 1997;112:1845–53.
Ahmad T, Marshall SE, Jewell D. Genetics of inflammatory bowel disease: the role of the HLA complex. World J Gastroenterol. 2006;12:3628–35.
Chervonsky AV. Influence of microbial environment on autoimmunity. Nat Immunol. 2010;11:28–35.
Birney E. Chromatin and heritability: how epigenetic studies can complement genetic approaches. Trends Genet. 2011;5:172–6.
Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;2:142–7.
Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology. 2013;145:293–308.
Acknowledgments
This work was supported in part by Health and Labor Sciences Research Grants for Research on Intractable Diseases from the Ministry of Health, Labor and Welfare of Japan (Y.A.). We are very grateful to Dr. Y Numata, Dr. H Nasuno, and Dr. Y Ishimine, residents of the First Department of Internal Medicine, for their resource collection.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arimura, Y., Isshiki, H., Onodera, K. et al. Characteristics of Japanese inflammatory bowel disease susceptibility loci. J Gastroenterol 49, 1217–1230 (2014). https://doi.org/10.1007/s00535-013-0866-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-013-0866-2