Abstract
Background
Chemotherapy-induced peripheral neuropathy (CIPN) is a common toxicity of taxanes for which there is no effective intervention. Genomic CIPN risk determination has yielded promising, but inconsistent results. The present study assessed the utility of a collective SNP cluster identified using novel analytics to describe taxane-associated CIPN risk.
Methods
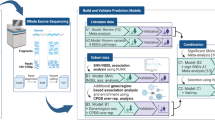
We analyzed GWAS data derived from ECOG-5103, first identifying SNPs that were most strongly associated with CIPN using Fisher’s ratio (FR). We then ranked ordered those SNPs which discriminated CIPN-positive (CIPN +) from CIPN-negative phenotypes based on their discriminatory power and developed the cluster of SNPs which provided the highest predictive accuracy using leave-one-out cross-validation (LOOCV).
Results
Using aggregated genotype data obtained from the previously reported ECOG-5103 clinical trial (in which two different arrays were used, HumanOmniExpress (727,227 SNPs) and HumanOmni1-Quad1 (1,131,857 SNPs)), we identified a 267 SNP cluster which was associated with a CIPN + phenotype with an accuracy of 96.1%.
Conclusions
A cluster of SNPs was identified which prospectively discriminated patients most likely to develop symptomatic CIPN following taxane exposure as part of a breast cancer chemotherapy regimen. Validation using an independent patient cohort should be performed.


Similar content being viewed by others
Abbreviations
- CIPN :
-
Chemotherapy-induced peripheral neuropathy
- GWAS :
-
Standard genome-wide association studies
- SNP:
-
Single-nucleotide polymorphisms
- HOQ1 :
-
HumanOmni1-Quad1
- HOE :
-
Holdout experiment
- LOOCV :
-
Leave-one-out cross-validation
- CDF :
-
Cumulative distribution function
References
Cavaletti G et al (2019) Chemotherapy-induced peripheral neurotoxicity: a multifaceted, still unsolved issue. J Peripher Nerv Syst 24(Suppl 2):S6–S12
Autissier E (2019) Chemotherapy-induced peripheral neuropathy: association with increased risk of falls and injuries. Clin J Oncol Nurs 23(4):405–410
Shah A et al (2018) Incidence and disease burden of chemotherapy-induced peripheral neuropathy in a population-based cohort. J Neurol Neurosurg Psychiatry 89(6):636–641
Hu S et al (2019) Recent developments of novel pharmacologic therapeutics for prevention of chemotherapy-induced peripheral neuropathy. Clin Cancer Res 25(21):6295–6301
Jordan B et al (2019) Prevention and management of chemotherapy-induced polyneuropathy. Breast Care (Basel) 14(2):79–84
Cliff J et al (2017) The molecular genetics of chemotherapy-induced peripheral neuropathy: a systematic review and meta-analysis. Crit Rev Oncol Hematol 120:127–140
Dolan ME et al (2017) Clinical and genome-wide analysis of cisplatin-induced peripheral neuropathy in survivors of adult-onset cancer. Clin Cancer Res 23(19):5757–5768
Leibovici A, Sharon R, Azoulay D (2018) BDNF Val66Met is associated with pre-existing but not with paclitaxel-induced peripheral neuropathy in an Israeli cohort of breast cancer patients. Isr Med Assoc J 20(12):746–748
Mahmoudpour SH et al (2018) Chemotherapy-induced peripheral neuropathy: evidence from genome-wide association studies and replication within multiple myeloma patients. BMC Cancer 18(1):820
Park SB et al (2017) Clinical and genetic predictors of paclitaxel neurotoxicity based on patient- versus clinician-reported incidence and severity of neurotoxicity in the ICON7 trial. Ann Oncol 28(11):2733–2740
Terrazzino S et al (2015) Genetic determinants of chronic oxaliplatin-induced peripheral neurotoxicity: a genome-wide study replication and meta-analysis. J Peripher Nerv Syst 20(1):15–23
Abraham JE et al (2014) Replication of genetic polymorphisms reported to be associated with taxane-related sensory neuropathy in patients with early breast cancer treated with Paclitaxel. Clin Cancer Res 20(9):2466–2475
Reyes-Gibby CC et al (2018) Genome-wide association study identifies genes associated with neuropathy in patients with head and neck cancer. Sci Rep 8(1):8789
Boora GK et al (2016) Testing of candidate single nucleotide variants associated with paclitaxel neuropathy in the trial NCCTG N08C1 (Alliance). Cancer Med 5(4):631–639
Marigorta UM et al (2018) Replicability and prediction: lessons and challenges from GWAS. Trends Genet 34(7):504–517
Zhou RH et al (2020) Co-expression gene modules involved in cisplatin-induced peripheral neuropathy according to sensitivity, status, and severity. J Peripher Nerv Syst 25(4):366–376
Hashemi S et al (2017) Exploring genetic attributions underlying radiotherapy-induced fatigue in prostate cancer patients. J Pain Symptom Manage 54(3):326–339
Reinbolt RE et al (2018) Genomic risk prediction of aromatase inhibitor-related arthralgia in patients with breast cancer using a novel machine-learning algorithm. Cancer Med 7(1):240–253
Sonis S et al (2013) SNP-based Bayesian networks can predict oral mucositis risk in autologous stem cell transplant recipients. Oral Dis 19(7):721–727
Schneider BP et al (2015) Genome-wide association studies for taxane-induced peripheral neuropathy in ECOG-5103 and ECOG-1199. Clin Cancer Res 21(22):5082–5091
Saligan LN et al (2014) Supervised classification by filter methods and recursive feature elimination predicts risk of radiotherapy-related fatigue in patients with prostate cancer. Cancer Inform 13:141–152
Fernández-Martínez JL et al (2018) Sampling defective pathways in phenotype prediction problems via the holdout sampler. Springer International Publishing, Cham
Liu F et al (2012) Performance comparison of multiple microarray platforms for gene expression profiling. Methods Mol Biol 802:141–155
Brewer JR et al (2016) Chemotherapy-induced peripheral neuropathy: current status and progress. Gynecol Oncol 140(1):176–183
Willson ML et al (2019) Taxanes for adjuvant treatment of early breast cancer. Cochrane Database Syst Rev 9:CD004421
Palmirotta R et al (2018) SNPs in predicting clinical efficacy and toxicity of chemotherapy: walking through the quicksand. Oncotarget 9(38):25355–25382
Tam V et al (2019) Benefits and limitations of genome-wide association studies. Nat Rev Genet 20(8):467–484
Chung, R.-H., C.-Y. Kang (2019) A multi-omics data simulator for complex disease studies and its application to evaluate multi-omics data analysis methods for disease classification. GigaScience 8(5).
Kivelä M et al (2014) Multilayer networks. J Complex Netw 2(3):203–271
Chan A et al (2019) Biological predictors of chemotherapy-induced peripheral neuropathy (CIPN): MASCC neurological complications working group overview. Support Care Cancer 27(10):3729–3737
Cavaletti G et al (2010) Chemotherapy-Induced Peripheral Neurotoxicity assessment: a critical revision of the currently available tools. Eur J Cancer 46(3):479–494
Tan e.a. (2019) Chemotherapy-induced peripheral neuropathy- patient reported outcomes compared with NCI CTCAE grade. Support Care Cancer.
Le-Rademacher e.a. (2017) Patient-reported (EORTC) QLQ-CIPN 20) versus physician-reported (CTCAE) quantification of oxaliplatin-and paclitoxel/carboplatin-induced peripheral neuropathy in NCCTG/Alliance Clinical trials.
Acknowledgements
We thank Dr. Antonio Wolff (Johns Hopkins University), Dr. Kathy Miller (Indiana University), Dr. George Sledge (Stanford University), and Anne O’Neill (Harvard/Dana Farber) and all the patients who participated in ECOG 5103 for providing access to the data that were used to analyze the reported results in this manuscript.
Funding
This work was supported by grants from the Division of Cancer Prevention, National Cancer Institute (R01CA238946-02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
SS and MBL conceived the study concept and wrote the manuscript and edited the manuscript. XW participated in the design of the study, data collection, and analysis, and drafted the manuscript. JLFM and E. J. d. A. G. participated in data analysis, algorithm, and interpretation. SP and BS participated in the design of the study; data analysis and implementation of machine learning algorithm interpretation. JL participated in data collection and data analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This is a secondary analysis of a completed clinical trial ECOG 5103. ECOG-ACRIN E5103 was previously approved, including additional secondary analyses of genomic data, by the Central Institutional Review Board (CIRB) for the National Cancer Institute (NCI) and conducted in accordance with Good Clinical Practice guidelines as defined by the International Conference on Harmonisation. All patients provided IRB-approved written informed consent. This study was monitored by the ECOG-ACRIN Data Safety Monitoring Committee and the NCI. The current secondary analysis has been approved by the ECOG Executive Board. The National Cancer Institute Central Institutional Review Board (IRB) approved the protocol the IRB of record for a subset of institutions; the remaining sites used their own individual IRBs. Written informed consent was obtained from all participants. Secondary analyses were in accordance with original consent and IRB approval and were further approved by the ECOG Executive Board.
Competing interests
Dr. Sonis reports personal fees from Biomodels, LLC, and personal fees from Primary Endpoint Solutions, LLC, outside of the submitted work. As an employee of Biomodels and PES, he is involved in assisting industry, government, and academics in the study and enablement of drugs, biologicals, and devices to treat patients for a broad range of indications including cancer and toxicities of cancer therapy. He does not have equity or receive payment from any of the companies’ clients. The other authors have no relevant disclosers.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lustberg, M., Wu, X., Fernández-Martínez, J.L. et al. Identification of a SNP cluster associated with taxane-induced peripheral neuropathy risk in patients being treated for breast cancer using GWAS data derived from a large cooperative group trial. Support Care Cancer 31, 139 (2023). https://doi.org/10.1007/s00520-023-07595-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00520-023-07595-9