Abstract
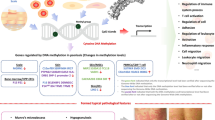
Epigenetics is the study of heritable changes in gene expression that do not originate from alternations in the DNA sequence. Epigenetic modifications include DNA methylation, histone modification, and gene silencing via the action of microRNAs. Epigenetic dysregulation has been implicated in many disease processes. In the field of dermatology, epigenetic regulation has been extensively explored as a pathologic mechanism in cutaneous T-cell lymphoma (CTCL), which has led to the successful development of epigenetic therapies for CTCL. In recent years, the potential role of epigenetic regulation in the pathogeneses of inflammatory skin diseases has gained greater appreciation. In particular, epigenetic changes in psoriasis and atopic dermatitis have been increasingly studied, with DNA methylation the most rigorously investigated to date. In this review, we provide an overview of DNA methylation in inflammatory skin diseases with an emphasis on psoriasis and atopic dermatitis.
Similar content being viewed by others
References
Waddington CH (2012) The epigenotype. Int J Epidemiol 41(1):10–13
Rodríguez-Paredes M, Esteller M (2011) Cancer epigenetics reaches mainstream oncology. Nat Med 17(3):330–339
Berger SL, Kouzarides T, Shiekhattar R et al (2009) An operational definition of epigenetics. Genes Dev 23(7):781–783
Feinberg AP, Koldobskiy MA, Göndör A (2016) Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17(5):284–299
Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150(1):12–27
Wu X, Zhang Y (2017) TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18(9):517–534
Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8(4):286–298
Feinberg (2018) The key role of epigenetics in human disease. N Engl J Med 379(4):400–401
Li Y, Sawalha AH, Lu Q (2009) Aberrant DNA methylation in skin diseases. J Dermatol Sci 54(3):143–149
Shanmugam MK, Sethi G (2013) Role of epigenetics in inflammation-associated diseases. Subcell Biochem 61:627–657
Jeltsch A, Jurkowska RZ (2014) New concepts in DNA methylation. Trends Biochem Sci 39(7):310–318
Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6(8):597–610
Boehncke WH, Schön MP (2015) Psoriasis. Lancet 386:983–994
Grozdev I, Korman N, Tsankov N (2014) Psoriasis as a systemic disease. Clin Dermatol 32(3):343–350
Gladman DD, Antoni C, Mease P et al (2005) Psoriatic arthritis: epidemiology, clinical features, course, and outcome. Ann Rheum Dis 64(Suppl 2:ii1):4–7
Lu Q (2013) The critical importance of epigenetics in autoimmunity. J Autoimmun 41:1–5
Wang Z, Long H, Chang C, Zhao M, Lu Q (2018) Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: a comprehensive overview. Cell Mol Life Sci 75(18):3353–3369
Zhao M, Lu Q (2018) The aberrant epigenetic modifications in the pathogenesis of psoriasis. J Investig Dermatol Symp Proc 19(2):S81–S82.
Pollock RA, Abji F, Gladman DD (2017) Epigenetics of psoriatic disease: A systematic review and critical appraisal. J Autoimmun. 78:29–38
Traupe H, van Gurp PJ, Happle R et al (1992) Psoriasis vulgaris, fetal growth, and genomic imprinting. Am J Med Genet 42(5):649–654
Generali E, Ceribelli A, Stazi MA et al (2017) Lessons learned from twins in autoimmune and chronic inflammatory diseases. J Autoimmun 83:51–61
Xiang Z, Yang Y, Chang C, Lu Q (2017) The epigenetic mechanism for discordance of autoimmunity in monozygotic twins. J Autoimmun 1(83):43–50
Zhang P, Su Y, Chen H et al (2010) Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J Dermatol Sci 60(1):40–42
Yooyongsatit S, Ruchusatsawat K, Noppakun N et al (2015) Patterns and functional roles of LINE-1 and Alu methylation in the keratinocyte from patients with psoriasis vulgaris. J Hum Genet 60(7):349–355
Murata Y, Bundo M, Ueda J et al (2017) DNA methylation and hydroxymethylation analyses of the active LINE-1 subfamilies in mice. Sci Rep 7(1):13624
Roberson ED, Liu Y, Ryan C et al (2012) A subset of methylated CpG sites differentiate psoriatic from normal skin. J Invest Dermatol. 132(3):583–592
Verma D, Ekman AK, Bivik Eding C et al (2018) Genome-wide DNA methylation profiling identifies differential methylation in uninvolved psoriatic epidermis. J Invest Dermatol 138(5):1088–1093
Zhang P, Zhao M, Liang G et al (2013) Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J Autoimmun 41:17–24
Zhou F, Wang W, Shen C et al (2016) Epigenome-wide association analysis identified nine skin DNA methylation loci for psoriasis. J Invest Dermatol 136(4):779–787
Tang L, Cheng Y, Zhu C et al (2018) Integrative methylome and transcriptome analysis to dissect key biological pathways for psoriasis in Chinese Han population. J Dermatol Sci 91(3):285–291
Chen M, Chen ZQ, Cui PG et al (2008) The methylation pattern of p16INK4a gene promoter in psoriatic epidermis and its clinical significance. Br J Dermatol 158(5):987–993
Chen M, Wang Y, Yao X et al (2016) Hypermethylation of HLA-C may be an epigenetic marker in psoriasis. J Dermatol Sci. 83(1):10–16
Chandra A, Senapati S, Roy S et al (2018) Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis. Clin Epigenetics 10(1):108
Gervin K, Vigeland MD, Mattingsdal M et al (2012) DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genes. PLoS Genet 8(1):e1002454
Han J, Park SG, Bae JB et al (2012) The characteristics of genome-wide DNA methylation in naïve CD4+ T cells of patients with psoriasis or atopic dermatitis. Biochem Biophys Res Commun 422(1):157–163
Park GT, Han J, Park SG et al (2014) DNA methylation analysis of CD4+ T cells in patients with psoriasis. Arch Dermatol Res. 306(3):259–268
Gu X, Nylander E, Coates PJ et al (2015) Correlation between reversal of DNA methylation and clinical symptoms in psoriatic epidermis following narrow-band UVB phototherapy. J Invest Dermatol 135(8):2077–2083
Bieber T (2008) Atopic dermatitis. N Engl J Med 358(14):1483–1494
Liang Y, Chang C, Lu Q (2016) The genetics and epigenetics of atopic dermatitis—filaggrin and other polymorphisms. Clin Rev Allergy Immunol 51(3):315–328
Lockett GA, Soto-Ramírez N, Ray MA et al (2016) Association of season of birth with DNA methylation and allergic disease. Allergy 71(9):1314–1324
Wang IJ, Chen SL, Lu TP et al (2013) Prenatal smoke exposure, DNA methylation, and childhood atopic dermatitis. Clin Exp Allergy 43(5):535–543
Nguyen CM, Liao W (2015) Genomic imprinting in psoriasis and atopic dermatitis: a review. J Dermatol Sci 80(2):89–93
Nakamura T, Sekigawa I, Ogasawara H et al (2006) Expression of DNMT-1 in patients with atopic dermatitis. Arch Dermatol Res 298(5):253–256
Novak N, Bieber T, Leung DY (2003) Immune mechanisms leading to atopic dermatitis. J Allergy Clin Immunol. 112(6 Suppl):S128–S139
Luo Y, Zhou B, Zhao M et al (2014) Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin Exp Dermatol 39(1):48–53
Ziyab AH, Karmaus W, Holloway JW et al (2013) DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol 27(3):e420–e423
Rodríguez E, Baurecht H, Wahn AF et al (2014) An integrated epigenetic and transcriptomic analysis reveals distinct tissue-specific patterns of DNA methylation associated with atopic dermatitis. J Invest Dermatol 134(7):1873–1883
Leoni C, Montagner S, Rinaldi A et al (2017) DNMT3a restrains mast cell inflammatory responses. Proc Natl Acad Sci USA 114(8):E1490–E1499
Leoni C, Montagner S, Deho' L et al (2015) Reduced DNA methylation and hydroxymethylation in patients with systemic mastocytosis. Eur J Haematol 95(6):566–575
Shukla A, Sehgal M, Singh TR (2015) Hydroxymethylation and its potential implication in DNA repair system: a review and future perspectives. Gene 564(2):109–118
Hessam S, Sand M, Lang K et al (2017) Altered global 5-hydroxymethylation status in hidradenitis suppurativa: support for an epigenetic background. Dermatology 233(2–3):129–135
Cruz AF, de Resende RG, de Lacerda JCT et al (2018) DNA methylation patterns of genes related to immune response in the different clinical forms of oral lichen planus. J Oral Pathol Med. 47(1):91–95
Mervis JS, McGee JS (2019) Epigenetic therapy and dermatologic disease: moving beyond CTCL. J Dermatolog Treat 30(1):68–73
Funding
The authors received no funding for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent not required for this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mervis, J.S., McGee, J.S. DNA methylation and inflammatory skin diseases. Arch Dermatol Res 312, 461–466 (2020). https://doi.org/10.1007/s00403-019-02005-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00403-019-02005-9