Abstract
Seed dormancy is an adaptive trait that improves survival of the next generation by optimizing the distribution of germination over time. The agricultural and forest industries rely on seeds that exhibit high rates of germination and vigorous, synchronous growth after germination; hence dormancy is sometimes considered an undesirable trait. The forest industry encounters problems with the pronounced dormancy of some conifer seeds, a feature that can lead to non-uniform germination and poor seedling vigor. In cereal crops, an optimum balance is most sought after; some dormancy at harvest is favored because it prevents germination of the physiologically mature grain in the head prior to harvest (that is, preharvest sprouting), a phenomenon that leads to considerable damage to grain quality and is especially prominent in cool moist environments. The sesquiterpene abscisic acid (ABA) regulates key events during seed formation, such as the deposition of storage reserves, prevention of precocious germination, acquisition of desiccation tolerance, and induction of primary dormancy. Its regulatory role is achieved in part by cross-talk with other hormones and their associated signaling networks, via mechanisms that are largely unknown. Quantitative genetics and functional genomics approaches will contribute to the elucidation of genes and proteins that control seed dormancy and germination, including components of the ABA signal transduction pathway. Dynamic changes in ABA biosynthesis and catabolism elicit hormone-signaling changes that affect downstream gene expression and thereby regulate critical checkpoints at the transitions from dormancy to germination and from germination to growth. Some of the recent developments in these areas are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Control of seed germination and growth is crucial to the survival of the next generation, and there are critical checkpoints at the transitions from dormancy to germination and from germination to growth. At dispersal, the quiescent mature seed, when it encounters favorable environmental conditions (which can include light of a given wavelength, sufficient water, optimal temperatures, and adequate oxygen), commences germination. Germination, sensu stricto includes those events commencing with imbibition or uptake of water by the quiescent dry seed and culminates with the elongation of the radicle (Bewley and Black 1994; Bewley 1997). Visible evidence of the completion of germination is usually protrusion of the radicle through the seed structures surrounding the embryo (such as the testa and endosperm, or megagametophyte). Some seeds fail to complete germination under seemingly favorable conditions, even though they are viable. These dormant seeds must be exposed to environmental cues, such as periods of warm-dry conditions (after-ripening), moist chilling, or even smoke, for dormancy to be terminated (Adkins and others 1986; Egerton-Warburton 1998) (Figure 1). Dormancy in nature is clearly an adaptive trait, because it improves survival by optimizing the distribution of germination over time. For example, a seed that germinates in the spring after remaining dormant throughout the winter has a greater chance of successful seedling establishment than one that germinates immediately after its dispersal in the fall.
The development of primary and secondary dormancy in seeds and factors influencing its inception, degree, and termination. From Bewley and Black (1994). With permission from Plenum Press.
Dormancy can also be induced in mature, already dispersed, nondormant seeds (known as induced or secondary dormancy) by environmental conditions that are unfavorable for germination, for example, anoxia, unsuitable temperature or illumination (Bewley 1997) (Figure 1). In induced dormancy, the seed loses its sensitivity to dormancy-breaking factors (for example, light, nitrate, or other factors, depending on the species) (Bewley and Black 1994).
The agricultural and forest industries rely on seeds that exhibit high rates of germination and vigorous, synchronous growth after germination; hence dormancy is usually considered an undesirable trait. In agriculture, extensive breeding programs have reduced the degree of dormancy and improved other traits of crops, particularly yield. However, an optimum balance is most desirable; in cereal crops, some dormancy at harvest is favored because it prevents the germination of the physiologically mature grain in the head prior to harvest (that is, preharvest sprouting), a phenomenon that is especially prominent in cool moist environments. Indeed the lack of harvest dormancy in many cultivars of wheat has led to serious worldwide economic losses caused by the adverse effects of pre-harvest-sprouted wheat grains on end-product quality (Bewley and Black 1994; Gubler and others 2005). Preharvest sprouting is not only of import to the cereal industry, but also to Canada’s canola industry in relation to fall seeding. Spring seeding often produces low seed yields in crops such as canola that are sensitive to heat and drought stress during flower and seed set. Because these early developmental stresses are avoided by the fall seeding of the crop, several positive outcomes occur, including higher yield, increased pod and seed size, and higher oil content (Kirkland and Johnson 1999). The major impediment to fall seeding on a significant portion of Canadian canola acreage is premature germination. In the forest industry, tree orchards have been established to meet the planting demands for several conifer species. Here breeding programs are complicated by the fact that the generation times for trees are exceedingly long and many species are characterized by a deeply manifested dormancy. Forest nursery operations must accommodate lengthy treatments to break dormancy of these seeds; the treatments can be months in duration, and often are not optimized. Moreover, forest tree seeds have an inherent high genetic variability which results in great heterogeneity in their behavior and, in particular, in their rate of germination, their quality, and their vigor (El-Kassaby and others 1993).
The sesquiterpene abscisic acid (ABA) regulates many processes during the plant life cycle, including key events during seed formation, such as the deposition of storage reserves, prevention of precocious germination, the acquisition of desiccation tolerance, and the induction of primary dormancy. Its regulatory role is achieved by mechanisms that are largely unknown, in part by cross-talk with other hormones and their associated signaling networks. Functional genomics approaches hold promise for elucidating genes and proteins that control seed dormancy and germination, including components of the ABA signal transduction pathway. A greater understanding of the relative contributions of ABA biosynthesis and catabolism to dormancy maintenance and its termination will emerge from detailed genetic and biochemical studies of the enzymes that effect dynamic changes in seed and embryo ABA contents during the dormancy-to-germination transition. The recent cloning of the genes encoding key enzymes for the biosynthesis and metabolism of ABA may lead to unique approaches for germination control. These avenues of pursuit will contribute to our understanding of how dynamic changes in ABA flux and hormone signaling affect downstream gene expression, and thereby regulate critical check points at the transitions from dormancy to germination and from germination to growth. Some of the recent developments in these areas are discussed.
ROLE OF ABA IN THE PROMOTION OF DEVELOPMENTAL PROCESSES, PREVENTION OF PRECOCIOUS GERMINATION, AND INDUCTION OF SEED DORMANCY
In general, primary dormancy is imposed during seed and embryo development so that varying degrees and types of dormancy may be exhibited by the seed during the later stages of development (Kermode 1990; Bewley and Black 1994; Leadem 1990; Hilhorst 1995). Dormancy is generally classed into two major types: embryo dormancy and coat-enhanced dormancy (Bewley and Black 1994). In embryo dormancy, it is the embryo that is dormant, and the embryonic axis will not elongate even if the embryo is excised from its enclosing seed tissues and placed on water. Embryo dormancy is common in the Rosaceae and other woody species, and in some grasses such as wild oat (Avena fatua). In coat-enhanced dormancy, the embryo, when isolated is capable of germination, but the intact seed is dormant; thus, it is one or more of the enclosing seed tissues (for example, endosperm, perisperm, megagametophyte, pericarp, and testa) that imposes the block to germination. Species exhibiting coat-enhanced dormancy include most cereals, conifers, and several dicots. Some degree of both types of dormancy can exist simultaneously or successively in some species (Bewley and Black 2004). The inhibitory nature of the enclosing seed tissues may be attributed to one or a combination of the following effects: (1) interference with water uptake or gas exchange, (2) mechanical restraint, and (3) biosynthesis/action of inhibitors in the embryo and seed tissues. All these factors contribute to the coat-enhanced dormancy mechanisms of conifer seeds (Ren and Kermode 1999, 2000; Feurtado and others, 2003, 2004; Terskikh and others 2004; Schmitz and others 2000, 2002).
Regardless of whether a seed is dormant or non-dormant at maturity, an important role of ABA during early seed development is to maintain embryos in a developmental mode until they are fully formed and have accumulated sufficient reserves to permit successful germination and subsequent seedling establishment. Thus, ABA biosynthesis in the embryo and surrounding seed tissues encourages the continuation of maturation processes, including the synthesis of storage proteins and storage lipids; it also suppresses precocious germination of embryos (reviewed in Kermode 1990, 1995). Seeds of different species show differences in the timing of dormancy inception during development; the role of ABA in this process is evident from genetic and physiological studies (reviewed in Bewley 1997; Koornneef and others 2002). Typically, the level of ABA in seeds is low during early development (that is, during the histodifferentiation and early pattern formation stage); it increases thereafter and usually peaks around mid-maturation. Abscisic acid levels usually decline precipitously during late development, particularly during the maturation drying phase (Bewley and Black 1994; Meinke 1995; Kermode 1995; Bewley 1997 and references therein; also see Taylor and others [2005]). These developmental changes in endogenous ABA correlate well with the pattern of expression of the gene encoding the ABA biosynthesis enzyme zeaxanthin epoxidase (reviewed in Seo and Koshiba 2002; see references therein). However, several factors add to the complexity of the pattern of ABA increase and decline, including developmental changes in the sensitivities of embryo and seed tissues to ABA and the differential thresholds for the initiation and maintenance of developmental events (for example, the expression of certain developmental genes and the induction and maintenance of dormancy) (Xu and Bewley 1991; Jiang and others 1996; Kermode 1995). Moreover, the environment during seed development (for example, light, temperature, and water availability) can strongly influence the ABA content and sensitivity of the mature seed. In wheat, low temperatures can effect changes in ABA content or sensitivity, which in turn influence the degree of dormancy during development and in the mature grain (Walker-Simmons 1990; Garello and LePage-Degivry 1999). Likewise, water stress imposed during the development of Sorghum bicolor seeds decreases both their ABA content and sensitivity, and the seeds have an increased capacity for germination during development (Benech-Arnold and others 1991). In the barley cultivar Triumph and its mutant line TL43, endogenous ABA contents during grain development alone do not adequately account for genetic differences in germination percentages within two environments—one conducive to low dormancy and the other promotive of high dormancy (in Spain and Scotland, respectively) (Romagosa and others 2001).
Often there is no clear relationship between the ABA content of the mature dry seed or grain and the degree of dormancy. Thus, although ABA is important for dormancy inception in developing seeds, high levels of ABA need not be present to maintain the state of dormancy (for example, during late maturation and desiccation). However, during the process of imbibition, dormant versus non-dormant seeds can be clearly differentiated. Seeds remaining dormant continue to synthesize ABA during imbibition; those seeds that have been subjected to an effective dormancy-breaking treatment (for example, after-ripened or moist-chilled seeds) still synthesize ABA, but generally, ABA catabolism is favored—that is, there has been a shift in the ratio of the capacity for ABA biosynthesis versus catabolism (see below, under Dormancy Termination is Accompanied by Changes in ABA Biosynthesis, Turnover, and Sensitivity).
Some mutants characterized by maternally inherited reduced seed dormancy exhibit altered testa characteristics (reviewed in Koornneef and others 2002; Debeaujon and others 2000, 2001). This indicates the importance of the testa structure as a constraint to radicle emergence; seeds weakened in testa structure or having reduced pigmentation exhibit reduced seed dormancy. In the case of reduced testa pigmentation, structural weakening may arise because of reduced anthocyanidins. Thus, it is the polymeric nature of proanthocyanidins and their ability to bind proteins that confers the cell-cementing properties to the testa and allow it to act as an impermeable barrier that contributes to the inhibition of water uptake and germination (Debeaujon and others 2001). In Arabidopsis, removal of the testa allows germination of both GA-deficient mutants (Debeaujon and Koornneef 2000) and accessions that have a very strong dormancy (Koornneef and others 2002). The abnormal fragility of the testa of dag1 mutants of Arabidopsis (Papi and others 2000) may allow them to germinate in response to much lower red light fluence rates, and may allow more rapid progression to the phytochrome-independent stage (Papi and others 2002).
A regulatory role of ABA in testa development requires further investigation (reviewed in Hilhorst 1995). Abscisic acid–deficient and ABA-insensitive mutants of Arabidopsis exhibit reduced thickness of the mucilage layer surrounding the testa (Karssen and others 1983), which in turn is expected to alter the rate of water and oxygen uptake. The ABA-deficient mutant of tomato (Lycopersicon esculentum) (sit w) produces seeds that have thinner testas because they are comprised of fewer cell layers (one cell layer, versus the 5-cell layer characteristic of the wild-type); the mutant seeds are less sensitive to highly negative osmotic potentials, perhaps as a consequence of their structure (Hilhorst and Downie 1994; Hilhorst 1995).
ABA SIGNALING PATHWAY COMPONENTS: PUTATIVE ROLES IN DEVELOPMENT AND DORMANCY INCEPTION
Elucidating the role of ABA in controlling seed and embryo maturation processes, including the inception of dormancy, has been facilitated by the generation and analysis of mutants (for example, of tomato, maize, and Arabidopsis) that are either deficient in ABA or that exhibit a relative insensitivity to ABA (so-called response mutants). These latter mutants have defects in ABA signaling pathways and are characterized by reduced dormancy, which is generally accompanied by disruption of seed maturation and precocious expression of germinative/postgerminative genes. Screens to identify suppressors of ABA signaling mutants have helped to define some of the components of ABA signaling that interact with other signaling pathways and thus form the basis of hormonal networks that control the dormancy-to-germination transition.
Dormancy of developing seeds is dependent on ABA that is synthesized in the embryo and not on maternal sources of ABA (Karssen and others 1983; Frey and others 2004). However, the surrounding seed tissues and parent plant may nonetheless play an important role in maintaining or encouraging ABA biosynthesis in developing embryos.
The various ABA mutants have been important in linking ABA to key developmental processes, including the acquisition of desiccation tolerance, prevention of vivipary, and induction of primary dormancy. Vivipary is characterized by germination of the embryo within the fruit on the parent plant. There is an uninterrupted progression from embryogenesis to germination, with little or no cessation of growth (quiescence) and, in most cases, little or no dehydration. Vivipary is a normal occurrence in Rhizophora mangle (the mangrove), in which seeds and seedlings exhibit a relative insensitivity to ABA (Sussex 1975). The ABA-deficient sitiens mutant of tomato has approximately 10% the ABA content of its wild-type counterpart. This mutant exhibits reduced dormancy, and its seeds germinate viviparously in overripe fruits (Groot 1987; Groot and others 1991). Low endogenous ABA may not directly contribute to vivipary in this case, however, because the embryos also exhibit reduced sensitivity to the highly negative osmotic potential of surrounding seed tissues. Abscisic acid–deficient mutants of maize, Arabidopsis, and tobacco (viviparous (vp)5 or vp14, aba1, and aba2, respectively), exhibit reduced dormancy (Arabidopsis and tobacco) or vivipary (maize) (Karssen and others 1983; Tan and others 1997; Frey and others 1999; White and others 2000). Severe mutants of Arabidopsis exhibiting relative ABA insensitivity—that is, some of the abi3 mutants, as well as double mutant aba/abi plants—produce seeds that are intolerant of desiccation (reviewed in Kermode 1995; see references therein). Seeds of the abi3-7 mutant exhibit reduced dormancy and sensitivity to ABA and have undetectable Em6 mRNA and protein and reduced transcripts for two 2S albumin proteins (Bies-Etheve and others 1999).
Evidence regarding the contribution of ABA biosynthesis and catabolism to dormancy status has come from transgenic studies. In tobacco, overexpression of the gene encoding the ABA biosynthesis enzyme zeaxanthin epoxidase (that is, ABA2) produces transgenic seeds with more pronounced dormancy than that of the wild-type; conversely, the downregulated expression of this gene by an antisense approach results in reduced levels of ABA and in reduced dormancy (Frey and others 1999). In the same vein, constitutive expression of a gene encoding a major enzyme of ABA catabolism (the 8′ hydroxylase gene CYP707A2) in transgenic Arabidopsis seeds (ecotype C24) leads to reduced amounts of ABA at seed maturity, and there is a lowered requirement for after-ripening of the seeds as compared to wild-type (that is, nontransgenic C24) seeds. Conversely, expression of a mutated CYP707A gene results in seeds requiring a longer after-ripening period to terminate dormancy (Millar and others, unpublished). Sequestration of ABA within developing transgenic tobacco seeds (effected by expression of an anti-ABA single-chain variable fragment antibody) leads to a marked disruption of storage reserve deposition. Reserve accumulation is reduced to the extent that the storage parenchyma cells of the seed more closely resemble plant vegetative cells (Phillips and others 1997). The transgenic seeds exhibit other features of seedlings, including green chloroplast–containing cotyledons, desiccation intolerance, and premature activation of the shoot apical meristem.
It is important to note that ABA synthesis in the ABA-deficient mutants (and in some transgenic plants), while decreased, is generally not completely abolished. Mutants are often “leaky,” allowing some biosynthesis of ABA, and redundant genes/pathways often exist to compensate for defective genes. Abscisic acid contents of the vp14 mutant of maize are about 30% that found in wild-type seeds (Tan and others 1997); hence, the ABA in these mutant seeds is sufficient to allow normal development to proceed (with respect to reserve deposition), and only the later maturation stages are altered (for example, vp14 seeds exhibit an enhanced capacity for precocious germination).
As intimated above, the phenotypes of the ABA-insensitive mutants are also variable, depending on the severity of the defective allele. Only the null alleles of the ABI3 gene, for example, lead to a failure to complete seed maturation, producing mutant seeds that are green, reduced in their storage reserve content, and desiccation-intolerant.
Characterization of the ABA-insensitive mutants of maize and Arabidopsis led to the cloning of genes involved in ABA signaling, some of which are transcription factors. To date six classes of transcription factors have been identified that appear to be essential for ABA- or seed-specific gene expression: ABI3/VP1, ABI4, ABI5, LEC1, LEC2, and FUS3 (reviewed in Finkelstein and others 2002). Lesions in the genes encoding these factors underlie some of the phenotypic characteristics of mutants that are disrupted in seed maturation. For example, the VP1 gene of cereals and the ABI3 gene of Arabidopsis encode transcription factors that regulate the expression of ABA-responsive genes during seed development (for example, storage-protein genes and LEA genes) (McCarty and others 1991; Giraudat and others 1992). The encoded proteins belong to the B3 domain family of transcription factors; all ABI3/VP1 proteins contain four conserved domains: an acidic activation domain and three basic domains, B1, B2, and B3. Since their isolation from these species, other ABI3/VP1 orthologs have been identified in many plant species, including rice (Oryza sativa), wheat (Triticum aestivum), carrot (Daucus carota), bean (Phaseolus vulgaris), the resurrection plant (Craterostigma plantagineum), poplar (Populus trichocarpa), and yellow-cedar (Chamaecyparis nootkatensis) (Lazarova and others 2002; reviewed in Zeng and others 2003; see references therein). Although unique features have been uncovered, there is generally a functional conservation among different ABI3/VP1 orthologs, as demonstrated by the successful complementation (rescue) of the severe Arabidopsis abi3 mutant (abi3-6) by trangenic expression of either the monocot VP1 gene (Suzuki and others 2001) or the conifer CnABI3 gene (Zeng and Kermode 2004).
Based primarily on transgenic and gene expression studies, the ABI3/VP1 family of proteins are thought to perform at least three functions within seeds:
-
1.
They mediate the action of ABA in promoting storage reserve accumulation and synthesis of desiccation/stress protectants (for example, oleosin, LEAs, peroxiredoxin-like proteins, and small heat shock proteins) (reviewed in Kermode and Finch-Savage 2002; Zeng and others 2003; Zeng and Kermode 2004; see references therein). ABI3 regulates seed storage-protein gene expression by acting synergistically with other transcription factors (for example, FUS3 and LEC1, LEC2, and others) that participate in combinatorial control (reviewed in Finkelstein and others 2002; Kroj and others 2003).
-
2.
They repress gene expression, particularly that of post-germinative genes (McCarty 1995; Nambara and others 1995, 2000; Paek and others 1998; Hoecker and others 1995, 1999). In some instances in which ABI3/VP1 genes are defective, the mutants’ seeds show an altered or premature activation of post-germinative gene expression (for example, genes encoding the chlorophyll a/b binding protein and isocitrate lyase in the Arabidopsis abi3 mutant and genes encoding malate synthase and isocitrate lyase in the maize vp1 mutant). VP1 also inhibits the induction of α-amylase gene expression in aleurone layer cells of developing maize seeds (Hoecker and others 1995); derepression of the expression of a chimeric α-amylase (5′)-GUS gene depends on the presence of a viviparous embryo (Hoecker and others 1999).
-
3.
They play a role in dormancy inception during development of seeds and may further maintain a dormant state in mature dispersed seeds. The ABI3 protein of yellow-cedar seeds may play a role in maintaining deep dormancy at seed maturity (Zeng and others 2003); likewise, the angiosperm counterparts of yellow-cedar, particularly the VP1 orthologs of cereals, have been implicated in the maintenance of embryo dormancy (see below, under Maintenance of Dormancy at Seed Maturity).
We can surmise that these putative functions of VP1 and ABI3 proteins are achieved by interactions with other proteins, especially transcription factors. In concert, these may effect activation or repression of target genes, including those important for maintaining/terminating the dormant state. ABI3/VP1 may recruit additional DNA-binding proteins to the promoters of storage-protein genes via its ability to alter chromatin structure (for example, nucleosome positioning) (Li and others 2001). Regulation of the expression of an Arabidopsis 2S storage protein gene (At2S3) appears to involve FUS3 and LEC2, both of which bind directly to promoter elements (RY repeats 1 and 2), and ABI3, which acts in an indirect manner (likely via its interaction with bZIP proteins that bind to the G-box) (Kroj and others 2003). ABI5 (a bZIP transcription factor) interacts directly via the B1 domain of ABI3 and two of the conserved charged domains of ABI5 that contain putative phosphorylation residues (Nakamura and others 2001). ABI5 binding to ABREs (ABA Responsive Elements) may tether ABI3 to target promoters and facilitate the interaction of ABI3 with RY elements (a consensus sequence conserved in many seed-specific gene promoters) and transcription complexes (Finkelstein and others 2002). The B2 domain of ABI3 is required for ABA-regulated gene expression and appears to facilitate the DNA binding capacity of a number of diverse DNA binding proteins (Carson and others 1997; Hill and others 1996). 14-3-3 proteins may facilitate interactions between VP1/ABI3 and other proteins by their ability to perform chaperone/scaffolding functions (Schultz and others 1998).
ABI3, together with ABI4 and ABI5, participates in combinatorial control of gene expression, possibly by forming a regulatory complex mediating seed-specific and/or ABA-inducible expression (Finkelstein and others 2002); there appears to be extensive cross-regulation of expression among ABI3, ABI4, and ABI5 genes (Söderman and others 2000). Mutations in the ABI4 and ABI5 gene loci have effects on seed development and ABA sensitivity that are similar to those associated with the abi3 mutant; however, null mutations in the ABI3 gene locus result in more severe phenotypes than those in the ABI4 or ABI5 gene loci (reviewed in Finkelstein and others 2002). The encoded proteins (ABI4 and ABI5) contain putative DNA binding and protein interaction domains. ABI4 is closely related to the APETALA2 domain family of transcription factors (within the AP2 domain) (reviewed in Finkelstein and others 2002). The ABI5 family of proteins all share three conserved charged domains (present in the N-terminal half of the protein and containing potential phosphorylation sites) and a bZIP domain at the C-terminus. Like VP1/ABI3, ABI5 can either activate or repress gene expression, and ABI3 and ABI5 may have antagonistic or synergistic effects on gene expression, depending on the gene (Finkelstein and Lynch 2000; Delseny and others 2001).
In comparison to the ABI5 gene, whose expression occurs late in the seed maturation program (that is, 17-20 DAP), ABI4 gene expression is detected from 8 DPA onward. It is highest in seeds where it is confined to the embryo, but there are some ABI4 gene transcripts present at low levels in vegetative tissues (Söderman and others 2000). Abi4 mutant seeds exhibit significantly reduced expression of LEA genes (for example, AtEm6 and PAP140). Likewise, abi5 mutant seeds exhibit reduced expression of certain LEA genes (AtEm1, AtEm6, and LeaD34), whereas the expression of some developmental genes is not altered (for example, the storage-protein gene vicilin and the gene encoding the integral oil body protein oleosin). ABI5 gene transcripts are downregulated in the abi1, abi2, abi3, abi4, and aba1 mutant backgrounds of Arabidopsis, suggesting that the ABI5 gene is regulated by ABA and these abi loci (Finkelstein and Lynch 2000).
Because ABA and ABA signaling play a key role in many developmental processes, it has been difficult to elucidate the most important components involved in dormancy inception. The role of ABA in controlling the transition from dormancy to germination likely involves actions at several levels—including effects on transcription, RNA processing, post-translational protein modifications, and the metabolism of secondary messengers. Some of these actions are discussed below, in the section on Other Components of ABA Signal Transduction.
MULTIPLE FACTORS FUNCTION IN CONCERT TO CONTROL SEED DEVELOPMENT
Along with ABI3, ABI4, and ABI5, other proteins act in a concerted fashion to promote events that are critical to the later stages of seed development, including the acquisition of desiccation tolerance and the induction of dormancy; at the same time, they appear to repress germinative and post-germinative functions (Holdsworth and others 1999). Arabidopsis LEAFY COTYLEDON (LEC) genes are defined by mutations at three loci, LEC1, LEC2, and FUSCA3 (FUS3). Recessive mutations at these gene loci lead to various abnormalities during early, mid, and late embryogenesis (reviewed in Harada 2001). Specification of cotyledon identity and maintenance of suspensor cell identity are early functions of the LEC genes (Meinke and others 1994; Lotan and others 1998). The later functions of the LEC genes are manifested in various defects, including a loss of dormancy and failure to acquire desiccation tolerance (for example, all known alleles of the lec1 mutation and strong fus3 mutant alleles). Synthesis and accumulation of storage proteins and lipids (and other gene products associated with reserve deposition) are substantially reduced in lec1 and fus3 embryos; interestingly, unlike wild-type embryos, the mutant embryos appear to compensate by accumulating starch grains (Keith and others 1994; Meinke and others 1994; Bäumlein and others 1994; Kirik and others 1996; Parcy and others 1997). In lec mutants, the rate of development is altered, and some characteristics of lec mutant embryos are seedling-like, indicating that postgerminative development is elicited precociously. For example, in Arabidopsis, the embryonic shoot apical meristems become activated during embryogenesis and more closely resemble postgerminative meristems. Thus, there is a defect in maturation processes that normally inhibit precocious germination and the expression of germinative and postgerminative genes (Harada 1997, 2001). Lec1 and fus3 mutant embryos occasionally exhibit vivipary.
Some of the effects of LEC gene defects on ABA biosynthesis and response have been examined. Lec1 mutant seeds are not sensitive to ABA (Parcy and others 1997; but see also Meinke and others 1994). Immature fus3 mutant siliques accumulate about one-third of the ABA level characteristic of wild-type seeds; however, by maturity, fus3 siliques have an equivalent amount of ABA to that of the wild type (Nambara and others 2000).
Both FUS3 and LEC1 encode putative transcription factors. Thus, they are thought to exert their effects by protein-DNA and/or protein–protein interactions. The LEC1 gene encodes a protein with sequence similarity to the HAP3 subunit of CCAAT binding factors. In mammalian cells, these binding factors, which are heterotrimeric, enhance the transcription of a large number of genes (reviewed in Harada 2001; see references therein). The pattern of expression of the Lec1 gene is not confined to the embryo, suggesting that its role is a global one—establishing an environment that promotes embryogenesis; this cellular environment likely coordinates the morphogenesis and maturation phases of development (Harada 2001).
Lec2 is able to establish a cellular environment sufficient to initiate embryo development; its ectopic expression during the post-embryonic phase induces the formation of somatic embryos and other organ-like structures and confers embryonic characteristics to seedlings (Stone and others 2001).
Fus3 mRNA accumulates primarily in developing siliques and is at maximal levels during mid-maturation (Leurssen and others 1998). FUS3 and LEC1 modulate the abundance of ABI3 protein in seeds, and synergistic interactions between the three proteins (ABI3, FUS3, and LEC1) are thought to control various key events, including accumulation of chlorophyll and anthocyanins, sensitivity to ABA, and expression of individual members of the 12S storage protein gene family (Parcy and others 1997). In transient transcription assays, FUS3 activates the transcription of maturation-specific genes containing the RY motif (Reidt and others 2000). Thus, both FUS3 and ABI3 may be essential components of a regulatory network acting in concert through the RY-promoter element to control gene expression during late embryogenesis and seed development. Notably, none of the ABI transcription factors (ABI3, ABI4, or ABI5) appear to interact directly with either FUS3 or LEC1 in a yeast two-hybrid assay (Brocard-Gifford and others 2003).
FUS3 and LEC2 also appear to control GA biosynthesis (see discussion in the following section). In the Arabidopsis mutants with defective expression of these genes (fus3 and lec2), the GA-pathway is misregulated, and this in turn leads to some characteristics of the altered phenotype (for example, trichome cell formation, a GA-regulated process) (Curuba and others 2004).
ROLE OF ABA–GIBBERELLIN INTERACTIONS IN DORMANCY AND GERMINATION
Abscisic acid and gibberellins (GAs) are generally believed to act antagonistically to control both dormancy breakage and germination (Karssen and Lacka 1986; Karssen 1995 and references therein). Although ABA controls seed developmental events, including the inception of dormancy, GAs are generally viewed as important for the promotion and maintenance of germination, for example, by promoting the growth potential of the embryo and by mediating the weakening of tissues that enclose the embryo (Bewley 1997). However, GA also appears to act as an antagonist to ABA function during seed development (White and others 2000). When an ABA-deficient mutant of maize (vp5) is manipulated either genetically or via biosynthesis inhibitors to induce GA deficiency during early seed development, vivipary is suppressed in developing kernels and the seeds acquire desiccation tolerance and storage longevity. In cultured immature maize embryos, GA deficiency induced by inhibiting biosynthesis enhances many ABA-responsive developmental events, including the accumulation of anthocyanins and transcripts encoding storage proteins and LEAs (White and Riven 2000). In situ, the major accumulation of GA1 and GA3 occurs in wild-type maize kernels, just prior to a peak in ABA content during development. It is speculated that these GAs induce a developmental program that leads to vivipary in the absence of normal amounts of ABA, and that a reduction of GAs re-establishes an ABA/GA ratio appropriate for suppression of germination and induction of maturation. Induction of GA deficiency does not suppress vivipary in vp1 mutant kernels, suggesting that VP1 acts downstream of both GA and ABA in programming seed development.
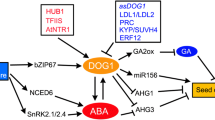
The embryonic regulators FUS3 and LEC2 that regulate multiple processes during seed development (for example, repression of postgerminative programs and activation of seed storage protein genes; see previous section) also appear to control GA biosynthesis. In the Arabidopsis mutants with defective expression of these genes (fus3 and lec2), the GA pathway is misregulated, and this misregulation leads to some characteristics of the altered phenotype (for example, trichome cell formation, a GA-regulated process; Curuba and others 2004). In wild-type seeds, the FUS3 protein represses expression of the gene encoding the GA biosynthetic enzyme AtGAox2 by physically interacting with two RY elements (GATGCATG) of the AtGAox2 promoter. Repression of the AtGAox2 gene by FUS3 occurs primarily in epidermal cells of the embryo axis, which is distinct from the pattern of expression of this gene during germination (Curuba and others 2004).
Expression of a GA 20-oxidase gene is different in the embryos of two lines of sorghum that have different levels of dormancy (Pérez-Flores and others 2003). In the low dormancy line, embryos within imbibed caryopses show detectable GA 20-oxidase gene transcripts during early imbibition (up to 18 h); expression of the gene increases thereafter, especially between 40 and 72 h. By contrast, in the high dormancy line, GA 20-oxidase gene transcripts remain at a low but detectable level throughout the 96-h imbibition period. (Note that only ungerminated seeds of both lines were used for these analyses). The expression of the sorghum GA 20-oxidase gene is enhanced in embryos of both lines following their excision from the caryopsis and incubation in water, illustrating that the enclosing seed tissues exert an inhibitory control over the expression of the gene. Abscisic acid may be the inhibitory factor in dormant seeds that represses GA biosynthesis, because exogenous ABA leads to a suppression of transcription of the gene in isolated embryos (Pérez-Flores and others 2003).
A subset of GA-regulated genes, whose expression is increased prior to radicle emergence in accordance with an increase in the levels of endogenous active GAs, has been characterized in Arabidopsis seeds. The GA-responsive genes activated include those responsible for the synthesis, transport, and signaling of other hormones, which underscores the importance of cross-talk during the germination process (Ogawa and others 2003). The site of expression of the GA-responsive genes is not localized to the site of GA biosynthesis, leading the authors to conclude that GA itself (or a GA signal) is transmitted across different cell types during Arabidopsis seed germination. The importance of hormonal cross-talk is also evident during dormancy termination of beech (Fagus sylvatica) seeds, in which application of inhibitors of ethylene or GA biosynthesis stimulates production of transcripts encoding GA20ox1 during moist chilling (Calvo and others 2004).
Dormancy in which exposure of seeds to chilling or light is required for its termination is often overcome by gibberellins (particularly GA3, GA4. and GA7), and synergistic interactions between phytochrome and applied GA have been noted. For example, skotodormancy is induced in seeds of Paulownia tomentosa by imbibing them in darkness for 30 days; however, this dormancy is readily overcome by exogenous GA3 or 6 h of red light (Grubisic and others 1985).
Reduced seed dormancy is exhibited by the rdo1 and rdo2 mutants of Arabidopsis (Leon-Kloosterziel and others 1996). Neither mutant is deficient in ABA biosynthesis, nor do the mutants exhibit any differences in their responsiveness to auxin, ethylene, or cytokinin as compared to wild-type Arabidopsis seed. However, the rdo2 mutant is less sensitive to the inhibition of germination by the GA-biosynthesis inhibitor tetcyclasis; thus a mutation in this gene somehow causes a reduced requirement for GA biosynthesis to terminate dormancy.
As already noted, it is widely accepted that GA promotes germination, and in support of this, mutations in loci that control GA biosynthesis and signal transduction can also affect germination potential. However, the mutants disrupted in GA response (for example, gai, sleepy1, spindly, rga, and pickle), are generally not seed-specific with respect to GA insensitivity (Peng and others 1997; Steber and others 1998; Jacobsen and others 1996; Silverstone and others 1998; Ogas and others 1997). The spindly gene product is a negative regulator of GA responses; interestingly, it has high homology to N-acetylglucosamine transferases, but establishing the putative roles of this specific protein modification in GA signaling remains elusive (Jacobsen and others 1996). The Arabidopsis mutant sleepy1 (sly1) was recovered as a suppressor of the abi1 mutation (Steber and others 1998); this mutant displays characteristics of a GA-response mutant (severe dwarfing at the vegetative stage, dark-green foliage, and underdeveloped petals and stamens), yet it cannot be rescued by exogenous GA. Mutation of the GA1 locus decreases germination and reduces the sensitivity of plant growth to GA (Derxk and others 1994).
The antagonistic effects of GA and ABA are well known in isolated barley aleurone layers in relation to control of the synthesis of α-amylase. GAMYB is a GA-regulated transcriptional activator of α-amylase gene expression in aleurone layer cells of germinating barley grains (see previous section). SLN1 (a product of the SLENDER or SLN1 gene locus and a negative regulator of GA signaling) is necessary for repression of GAMYB in barley aleurone cells (Gubler and others 2002). Interestingly, the activity of SNL1 in aleurone layer cells is regulated posttranslationally, and the level of this protein declines rapidly in response to GA prior to any increase in GAMYB. Giberellin stimulates the degradation of SLN1; ABA downregulates the expression of GAMYB but has no effect on the stability of SLN1. The action of ABA in blocking GA signaling then is downstream of SLN1 (Gubler and others 2002). The barley SLENDER gene appears to be the functional ortholog of GAI/RGA genes in Arabidopsis (Richards and others 2001). As components of the GA signaling pathway, GAI and RGA genes mediate GA regulation of stem elongation (Lee and others 2002). Also, RGL1 and RGL2 genes encode proteins that are closely related to GAI and RGA. Of the four proteins that belong to the DELLA subfamily of GRAS regulatory proteins, only RGL2 regulates seed germination in response to GA, and it acts as a negative regulator. In wild-type and mutant ga1-3 Arabidopsis seeds, transcripts encoding this gene increase in amount during a moist chilling period (4°C) and are maintained at high levels for as long as the seeds are kept at 4°C. Wild-type seeds immediately imbibed in water at 23°C also show an increase in RGL2 transcripts, with expression restricted to the elongating regions of pre-emergent and recently emerged radicles. Upon transfer of wild-type seeds from moist chilling conditions to 23°C, the RGL2-related transcripts decline, particularly upon completion of germination. In contrast, in non-germinating mutant ga1-3 seeds maintained at 23°C, RGL2 transcript levels remain high. There has been speculation that RGL2 functions to prevent germination upon imbibition; GA can overcome this inhibition and promote germination by downregulating RGL2 gene expression; it may further integrate environmental and internal cues to control germination (Lee and others 2002). Most recently it has been shown that RGL2 is rapidly degraded by GA treatment; the F-box protein SLEEPY1 (SLY1) is required for degradation of RGL2 (Tyler and others 2004). SLY1, a positive regulator of the GA signaling pathway in Arabidopsis, is likely a component of the ubiquitin E3 ligase SCF complex (McGinnis and others 2003). In response to the GA signal, then, SCFSLY1 may target proteins such as RGL2 for ubiquitination and subsequent degradation by the 26S proteosome. Interestingly, DELLA proteins appear to mediate ethylene, auxin, and GA responses and are suggested to play an integrative role in the phytohormone signal response network (Achard and others 2003).
MAINTENANCE OF DORMANCY AT SEED MATURITY
In tobacco seeds, the proportion of genes repressed or induced during dormancy maintenance does not appear to differ significantly in the general categories of primary metabolism, cellular communication/signaling, and transcriptional control (Bove and others 2005). Indeed, within dormant seeds, the upregulated genes belong to the general categories of cell rescue, secondary metabolism, and transport facilitation. Interestingly, upon dormancy termination, virtually all of the upregulated genes belong to the functional categories of cellular organization, regulated proteolysis (for example, photomorphogenesis repressor-like proteins of light signaling), and translational control. Thus, light perception, components of signaling networks, translational control, and regulated proteolysis are all linked to the maintenance of dormancy and its termination (Bove and others 2005).
Role of VP1/ABI3
Although there are clearly other transcriptional factors and components of ABA signaling that are involved in dormancy maintenance, the VP1/ABI3 gene product has perhaps received the most attention as a putative regulator of dormancy in mature seeds (Jones and others 1997; Fukuhara and others 1999; Nakamura and Toyama 2001; Carrari and others 2001; Zeng and others 2003). In Avena fatua (wild oat), expression of the VP1 gene is correlated with the degree of embryo dormancy and may be important for maintaining ABA-controlled metabolism in the imbibed seed (Jones and others 1997). For example, in embryos of A. fatua seeds, transcripts are higher in seeds that are stored at 4°C and are still dormant, but they decline in seeds that are fully after-ripened at 24°C (Jones and others 1997, 2000). A VP1 gene ortholog cloned from sorghum (Sorghum bicolor) has been examined in relation to its potential involvement in pre-harvest sprouting resistance (Carrari and others 2001). In pre-harvest sprouting, there is germination of the physiologically mature grain while it is still on the parent plant; this phenomenon is particularly prevalent when maturation of the grain takes place under low temperature and high-humidity conditions (Bewley and Black 1994). In wheat, barley, and sorghum cultivars that are susceptible to pre-harvest sprouting, there is a relative insensitivity of the embryo to ABA as compared to resistant cultivars, generally in the absence of any substantial differences in ABA content, or a higher ABA content in the susceptible cultivars (Walker-Simmons 1987; Steinbach and others 1995; Benech-Arnold and others 1999).
Interestingly, the expression pattern of the sorghum VP1 gene is different in two cultivars/genotypes exhibiting differential resistance to pre-harvest sprouting at physiological maturity. More specifically, in the embryos of the susceptible cultivar, transcripts encoding VP1 peak at a relatively early stage of grain development (20 days after pollination), whereas the peak in the resistant cultivar occurs at much later developmental stages, when seed maturation is almost complete (Carrari and others 2001).
The role of ABI3/VP1 proteins in the maintenance of dormancy of imbibed mature seeds needs to be more thoroughly addressed in a wider range of species and studies are clearly needed that examine expression at both the mRNA and protein levels. Maintenance of dormancy in the conifer species yellow-cedar, whose seeds are deeply dormant at maturity, may involve ABI3 (Zeng and others 2003). The protein (CnABI3) is present in the megagametophyte and embryo of dormant mature and warm stratified yellow-cedar seeds, but declines during subsequent moist chilling, a treatment effective in breaking dormancy. In contrast, the protein is preserved (albeit in lower amounts) in seeds subjected to a control treatment (12 weeks in warm moist conditions) that is ineffective in breaking dormancy. A decline in CnABI3 gene transcripts is also positively correlated with dormancy breakage; it does not occur during moist chilling itself, but rather during subsequent germination. Regulation of the CnABI3 gene in relation to dormancy control is likely not restricted to the transcriptional level and may additionally occur at the level of stability of the CnABI3 protein or involve some other post-translational change (Zeng and others 2003).
A two-hybrid screen in yeast has identified three proteins that interact specifically with VP1 of wild oat (Jones and others 2000); these proteins (referred to as AfVIP1, AfVIP2, and AfVIP3) may play specific roles in AfVP1-mediated regulation of the dormancy to germination transition. The AfVIP1 and AfVIP2 genes (encoding a ring-type zinc finger domain protein) are more highly expressed in dormant embryos, whereas expression of the AfVIP3 gene (encoding a paralog of the human C1 transcription factor involved in cell cycle control) is greater in germinated seeds (Jones and others 2000). Yeast two-hybrid screens using B2 and B3 domains of the Arabidopsis ABI3 protein as bait identified four putative interacting proteins: a CONSTANS-related factor, a protein related to the Drosophila transcription factor GOLIATH (involved in mesoderm formation), and the RPB5 subunit of RNA polymerase II (Kurup and others 2000). Yeast two-hybrid screens using the intact CnABI3 protein of yellow-cedar as bait identified three putative interacting proteins, including a heat shock protein binding factor (Zeng and Kermode unpublished). Further characterization of the various VP1/ABI3-interacting proteins uncovered by yeast two-hybrid screens, including the implementation of reverse genetics approaches, will aid in elucidating their specific roles in relation to the maintenance of deep dormancy.
Studies have mapped candidate genes such as VP1 to determine their association with dormancy QTLs. VP1 genes from wheat (Osa and others 2003) and sorghum (Carrari and others 2003) did not, however, correspond to dormancy QTLs. However, the finding that wheat VP1 transcripts are commonly mis-spliced, may indicate that their function is compromised, and this may underlie the low dormancy of many modern wheats (McKibbin and others 2002). The prospect that dormancy of wheat could be enhanced (and pre-harvest sprouting reduced) by expression of a VP1 gene orthog of wild oat has been noted (Gubler and others 2005).
Does ABA Prevent Cell Wall HydrolaseProduction?
For some seeds, embryos that are constrained by a mechanical barrier, such as the surrounding endosperm, perisperm, or megagametophyte (that is, those that exhibit coat-enhanced dormancy), appear to require a weakening of these structures to permit radicle protrusion. More specifically, a weakening of the cell walls of the surrounding seed tissues, especially at the micropylar region is proposed to be a prerequisite for germination. This process, mediated by cell wall hydrolases, would decrease the force required by the radicle to penetrate through seed tissues.
Hydrolytic enzymes implicated in dormancy termination include oxalate oxidase (Grzelezak and others 1985), endo-β-mannanase (Downie and others 1997), and pectin methyl esterase (Ren and Kermode 2000). In tomato seeds, chemical weakening of the surrounding endosperm is caused by enzymes produced under the influence of the embryo: dormant seeds are unable to produce the cell wall degrading enzymes. In these seeds, the endosperm cell walls contain relatively large amounts of galactomannans (Groot and others 1988), which are a carbohydrate reserve. Three enzymes contribute to the hydrolysis of the galactomannans: α-galactosidase, a mannohydrolase, and endo-β-mannanase. In germinating wild-type tomato seeds, lowering of the puncture force required by the radicle to penetrate the mannan-rich endosperm cell walls has been attributed to endo-β-mannanase, an enzyme produced within the endosperm itself at about the time of radicle emergence (Groot and others 1988; reviewed in Bewley 1997; see references therein). The enzyme is first produced in the micropylar region and later, after germination, in the rest of the endosperm; each region produces different isoforms. Subsequently it was revealed that only the isoforms of the enzyme that are produced following germination—the post-germinative forms (involved in reserve mobilization)—are potentially regulated by ABA. It has further been questioned whether endo-β-mannanase is in itself sufficient for germination to be completed, because the enzyme can accumulate in the micropylar region of the endosperm in the absence of germination, for example when the embryo growth potential is not limiting (reviewed in Bewley 1997). In coffee (Coffea arabica) seeds, ABA inhibits endosperm cap weakening, potentially by inhibiting the activities of at least two endo-β-mannanase isoforms and/or indirectly, by inhibiting the increase in the pressure force of the radicle (Amaral da Silva and others 2004).
Tobacco endosperm cell walls may be weakened by β-1,3-glucanase to permit germination, because the activity of this enzyme increases within the micropylar endosperm before radicle emergence; ABA retards both enzyme accumulation and endosperm rupture (Leubner-Metzger and others 1995). The involvement of this enzyme in germination is also supported by studies of its expression in transgenic plants. Expression of a sense gene construct, driven by an ABA-inducible promoter leads to enhanced activity of the β-1,3-glucanase and to increased endosperm rupture (Leubner-Metzger and Meins 2000).
The pattern of increase in pectin methyl esterase activities in yellow-cedar seeds coincides with the process of dormancy breakage, and the amount of enzyme activity produced is strongly correlated with the ability of seeds to germinate (Ren and Kermode 2000). Furthermore, in the absence of effective dormancy-breaking treatments (that is, when mature seeds are subjected to a 3-day soak or to a control treatment comprised of a 90-day period in warm, moist conditions), no enzyme activity is induced. Abscisic acid causes a significant suppression of the pectin methyl esterase isoforms associated with dormancy breakage and germination/growth in embryos and megagametophytes of intact seeds. GA3 has a promotive effect on pectin methyl esterase activities in the embryo and megagametophyte at the germinative stage. However, the role of endogenous gibberellins in dormancy breakage of yellow-cedar seeds is by no means clear.
OTHER COMPONENTS OF ABA SIGNALTRANSDUCTION
The role of ABA in controlling the transition from dormancy to germination likely involves actions at several levels, including effects on transcription, RNA processing, post-translational protein modifications, and the metabolism of secondary messengers. Thus, as well as transcription factors, many of the components of ABA signal transduction are protein-modifying enzymes, RNA-binding/processing proteins, GTP-binding proteins, enzymes of phospholipid or phosphoinositide metabolism, and proteins regulating vesicle trafficking or subcellular localization of proteins. Only selected examples of these will be discussed.
ABA Reception
As with any classical signal transduction pathway, ABA signaling is presumed to start with the reception of the hormone signal by a specific receptor. Interaction between the hormone and its receptor(s) then triggers a signal transduction cascade involving an array of intermediary proteins and secondary signals. The end point of the process is the upregulation or downregulation of expression of certain target genes (for example, those involved in the maintenance or termination of dormancy). However, the concept of a simple (linear) signal transduction chain has been replaced by the more realistic view that signaling networks involving interacting components likely control complex processes, including the dormancy-to-germination transition and the germination-to-growth transition. Thus, regulation of seed developmental processes involves considerable cross-talk between different hormone signaling pathways, and gene expression may be controlled by parallel pathways involving both ABA-dependent and ABA-independent steps.
There is evidence that ABA perception occurs both intracellularly and at the extracellular face of the plasma membrane. Purification of ABA-binding proteins (for example, from broad bean leaf epidermal cells; Zhang and others 2002) is a major step toward isolating ABA receptors. ABAP1, is an ABA-inducible protein associated with the plasma membrane of barley aleurone layer cells (Razem and others 2004); ABAP1 genes are present in many diverse monocot and dicot species. The recombinant protein exhibits high binding affinity for ABA, and it binds 3H-(+)-ABA most effectively at neutral pH. Furthermore, binding exhibits saturation kinetics and is specific for the natural enantiomer of ABA, S-(+)-ABA. The protein is also capable of binding to two precursors of ABA, (+)-ABA aldehyde and (+)-ABA alcohol, both of which are able to displace 3H-ABA, because of the lack of a strict structural requirement at the C1 position of the ABA molecule (Razem and others 2004).
A leucine-rich repeat receptor-like kinase (LRR-RLK1) appears to be involved in early ABA perception in Arabidopsis (Osakabe and others 2005). These types of plasma-membrane–associated proteins contain an extracellular leucine-rich repeat and a Ser/Thr kinase domain; in many cases, the leucine-rich repeat domain, or the protein complex, functions as a receptor of both peptide and non-peptide hormones (Osakabe and others 2005 and references therein). As well as being expressed in seedlings in response to ABA, the gene encoding the kinase is also expressed in embryos of mature Arabidopsis seeds. PRPK1 knock-out mutants and antisense-PRPK1 transgenic lines exhibit ABA insensitivity with respect to seed germination, plant growth, stomatal closure, and gene expression. The capacity of the protein to bind to ABA is being analyzed. In addition, plant receptor–like kinases may form dimers or tetramers to become active and transduce the ligand-binding signal to downstream cascades. Thus, the isolation and characterization of proteins that are specific partners of the putative receptor are key endeavors, because together they form the ABA signaling complex (Osakabe and others 2005).
Heterotrimeric G proteins and G protein–coupled receptors are early components of signal transduction pathways that control growth and differentiation in eukaryotes. Activation of G protein–coupled receptors as a result of the binding of an extracellular ligand triggers a transduction cascade that is mediated by a G protein; GTP binds to the α-subunit of the G protein, causing a conformational change that allows the α-subunit to dissociate from the βγ-subunit. Activation of the G protein in this manner then activates downstream components (ion channels and adenylate cyclase) that lead to increases in intracellular mediators (Ca2+ and cAMP, respectively). Ultimately, changes in cellular functions including the activation/repression of genes are effected. This particular pathway has not diversified over the course of plant evolution nearly to the extent of that in animals (Colucci and others 2002 and references therein). Recently a gene of Arabidopsis encoding a putative G protein–coupled receptor (that is, CGR1) was isolated. Overexpression of this gene in Arabidopsis creates transgenic plants that have an early flowering phenotype and produce seeds that lack dormancy. The transformants behave as if they have altered sensitivity to endogenous ABA and/or GA, and seeds are less sensitive to exogenous ABA than are wild-type seeds as far as their germination is concerned. Overexpression of the CGR1 gene also induces the expression of the MYB65 gene, an Arabidopsis homolog of the GAMYB gene of barley, whose encoded product regulates GA-mediated α-amylase induction in aleurone layer cells (Colucci and others 2002).
ABA Transcription Factors
As already noted, characterization of mutants that have altered sensitivity to ABA and the cloning of the wild-type genes involved in the underlying processes that are disrupted in the mutants (for example, resulting in enhanced or reduced dormancy) have led to the identification of certain components of the ABA signaling pathway. Many are transcription factors and hence are likely part of the later stages of signal transduction. Expression of the Mediator of ABA-Regulated Dormancy1 (MARD1) gene, encoding a zinc-finger protein, is upregulated by ABA in imbibed Arabidopsis seeds. Seeds of mutant plants with a T-DNA insertion in the MARD1 gene promoter region downstream of two ABREs germinate faster and are slightly less responsive to exogenous ABA than are wild-type seeds (He and Gan 2004).
A protein that interacts with the bZIP ABA-Response-Element Binding protein (AREB1) of barley, ARIA, an Arabidopsis arm repeat protein, is a positive ABA signaling component involved in regulating germination, seedling growth, and glucose/ABA/stress responses (Kim and others 2004). A nuclear transcription factor of the creosote bush (Larrea tridentate), a xerophytic species, is able to transactivate the promoter of the ABA-inducible gene HVA22 via its C-terminal WRKY domain and an N-terminal motif that is essential for the repression of some regulators of ethylene signaling. In doing so, the WRKY21 transcription factor interacts synergistically with ABA, VP1, and ABI5, and a complex of the three proteins is thought to function downstream of ABI1 (Zou and others 2004).
RNA Transport/Processing Proteins
RNA processing is a modulator of ABA signaling (reviewed in Kuhn and Schroeder 2003). For example, some ABA sensitivity mutants have defects in RNA-binding proteins, and it is thought that by influencing transcript abundance, RNA-binding proteins modulate ABA signaling. The various post-transcriptional controls appear to be effected through the alteration of mRNA processing events including splicing, 3′ end processing, nuclear export, transcript stability, and RNA degradation. A reporter system (the luciferase gene-coding region linked to an ABA-inducible promoter) was exploited to identify mutations resulting in increased ABA sensitivity—by identifying plants that showed abnormal bioluminescence in response to exogenous ABA (Xiong and others 2001). A further characterization of one of the mutants uncovered (sad1) indicated that it is indeed supersensitive to ABA (although it also displays reduced ABA biosynthesis), and one manifestation of this supersensitivity is an increased ability of ABA to inhibit seed germination and root growth and to enhance expression of some ABA-responsive genes (particularly ABA-dependent phosphatase type 2C genes, previously implicated as negative regulators of ABA signaling; Sheen 1998). Interestingly, the SAD1 gene encodes a protein with similarity to the Sm-like proteins that are structurally related to Sm proteins contained in snRNPs that function in RNA splicing (reviewed in McCourt 2001). Sm proteins (for example, of animals and yeast) are multifunctional; most of their functions are related to the control of RNA processing (splicing, nuclear export and degradation). Another essential modulator of ABA signaling is ABH1, an Arabidopsis homolog of a nuclear mRNA cap-binding protein (Hugouvieux and others 2001). Analyses of era1-2 and abh1 double mutants (both characterized by ABA hypersensitivity), suggest that ERA1 and ABH1 do not modulate the same negative regulator in ABA signaling (Hugouvieux and others 2002). Some of the transcripts whose abundance is reduced in the abh1 mutant are implicated in ABA signaling, including those encoding a putative negative regulator of ABA signaling, AtPP2C, the RESPONSIVE TO DEHYDRATION20 protein, and various proteins that regulate oxidative stress (Sheen 1998; Hugouvieux and others 2001). These and other studies intimate that RNA metabolism (for example, mRNA turnover) is essential for the modulation of ABA signaling and may in fact directly modulate ABA receptor function (McCourt 2001).
Post-translational Protein Modification
Other proteins that have been identified—either as positive or negative modulators of ABA signaling—are kinases, phosphatases, and farnesyl transferases, enzymes that are capable of effecting post-translational modifications of proteins. The targets of these modifying enzymes likely include proteins that act as intermediaries in signal transduction cascades. Changes in the phosphorylation status of a protein can in turn modulate its function and stability or half-life; farnesylation of proteins is also a key modulator of protein function because it allows proteins to insert into membranes.
Evidence that reversible protein phosphorylation plays a role in ABA signaling has come in part from the analyses of the abi1 and abi2 mutants of Arabidopsis, which lead to relative ABA insensitivity of seeds as well as of vegetative tissues (Koornneef and others 1984; Finkelstein and Somerville 1990). The ABI1 gene of Arabidopsis, encodes a member of the 2C class of serine/threonine protein phosphatases (PP2Cs), and its activity is regulated by proton and Mg2+ ions, but not by Ca2+ ions (Leube and others 1998). The ABI2 gene of Arabidopsis also encodes a 2C class protein phosphatase, and it is likely that a family of these phosphatases acts as redundant negative regulators of ABA signaling (Gosti and others 1999), a contention supported by analysis of the Arabidopsis genome. There are about 70 proteins with homology to members of the family in Arabidopsis, several of which are related to ABI1 and ABI2 proteins (The Arabidopsis Genome Initiative 2000). In each of the respective mutants (that is, abi1 and abi2), the gene encoding the PP2C has a point mutation that results in an amino acid substitution in the catalytic domain, thus diminishing the activity of the enzyme (Leung and others 1994, 1997; Meyer and others 1994; Rodriguez and others 1998); the mutant alleles of these genes are dominant. The mutant seeds exhibit reduced dormancy and leaves of the mutant plants are prone to wilting.
Even though the abi1 mutant is characterized by reduced sensitivity to ABA, ABI1 is indeed a negative regulator of ABA signaling (Gosti and others 1999). Protein phosphatase 2C activity is absent in intragenic revertants of abi1 and abi2 mutants, and the revertants are characterized by ABA hypersensitivity (Gosti and others 1999; Merlot and others 2001). Overexpression of ABI1 in maize protoplasts decreases the ABA-mediated activation of a chimeric gene driven by an ABA-inducible promoter and this, along with other evidence (Tahtiharju and Palva 2001), is consistent with the role of ABI1 as a negative regulator of ABA signaling (Sheen 1998).
Expression of the ATHB6 gene is upregulated by ABA and drought stress (Söderman and others 1999). This gene encodes a homeodomain transcription factor of the HD-Zip class; the protein interacts with ABI1 and is likely a key regulator of ABA signaling (Himmelbach and others 2002). The ATHB6 protein contains a phosphatase domain and a DNA-binding site at its N-terminus; both are required for its interaction with ABI1. The transcription factor acts as a negative regulator in ABA signaling and acts downstream of ABI1; its constitutive expression in transgenic Arabidopsis plants leads to ABA insensitivity in a subset of ABI1-dependent responses, one of which is manifested by a threefold to fivefold reduction in ABA responsiveness as far as germination is concerned (Himmelbach and others 2002).
A gene encoding a PP2C has been isolated from beechnut (Fagus sylvatica) (designated FsPP2C1). Expression of the gene is upregulated by ABA. Further, FsPP2C1 gene transcripts increase during a 2-week imbibition of dormant seeds, and they decline throughout a dormancy-breaking moist chilling treatment, especially after 6 weeks of moist chilling, a treatment that elicits approximately 50% germination (Lorenzo and others 2001).
In addition to PP2Cs that act as negative regulators of ABA signaling, there also appear to be phosphatases (PP2As) that function early in ABA signal transduction and act as positive regulators (Kwak and others 2002). One such protein is RCN1, and rcn1 mutant seeds are characterized by reduced ABA inhibition of germination and a reduction in the ABA-induced transcripts encoding RD29A.
Characterization of the protein kinase PKABA1 provides further support for a role of reversible protein phosphorylation in ABA signaling (Anderberg and Walker-Simmons 1992). PKABA1 is an ABA-responsive protein kinase whose synthesis is upregulated in the dormant mature-imbibed cereal grain. Transient expression of PKABA1 in cereal aleurone layer cells is able to suppress the GA-induced expression of postgerminative genes (those encoding α-amylases and a cysteine protease) (Gomez-Cadenas and others 1999). However, the antagonistic effect of ABA on GA-mediated induction of α-amylase is not altered by inhibiting PKABA1 gene expression by use of double-stranded RNA interference technology (that is, transient expression of a PKABA1 RNAi). Thus, two independent ABA signaling pathways may lead to the suppression of α-amylase in barley aleurone layer cells, one dependent and the other independent of PKABA1 (Zentella and others 2002).
What are some of the proteins subject to phosphorylation by the wheat kinase? A wheat protein (TaABF, a homolog of ABI5) binds to PKABA1 in yeast two-hybrid assays. The nature of this interaction is specific; that is, TaABF is a phosphorylation substrate of the kinase, and interactions do not occur with a mutant form of PKABA1 that lacks the nucleotide binding domain. Furthermore, PKABA1 synthesized in transgenic cell lines is able to phosphorylate synthetic peptides representing three specific regions of TaABF (Johnson and others 2002). PKABA1 gene expression is not exclusive to the seed; it is also responsive to exogenous ABA and to stress within vegetative tissues. In contrast, TaABF gene expression is seed-specific. It is suggested that TaABF serves as a physiological substrate for PKABA1 in the ABA signal transduction pathway during grain maturation, dormancy maintenance, and ABA-suppressed gene expression (Johnson and others 2002).
Disruption of ABA signal transduction can lead to enhanced seed dormancy; examples are the era (enhanced responsiveness to ABA) mutants, which exhibit both increased sensitivity to ABA and enhanced primary dormancy (Cutler and others 1996). The ERA1 locus encodes a β-subunit of a farnesyl transferase (Cutler and others 1996). Farnesyl transferases are key enzymes in signal transduction as they effect a post-translational modification (attachment of a 15-C farnesyl pyrophosphate to proteins containing a CaaX motif). Farnesylation and other lipid modifications (for example, geranylgeranylation, myristoylation, and palmitoylation) mediate the localization of proteins to membranes (Yang 2002). Membrane localization, in turn, is a prerequisite for the correct functioning of several signal transduction proteins, including those belonging to the small GTPase superfamily. It is quite possible that negative protein regulators of ABA signaling require farnesylation to function (for example, to attenuate ABA receptor activation).
Some evidence for this has come from an examination of ROP10, a member of the Arabidopsis Rop subfamily of RHO GTPases implicated in ABA signaling (Zheng and others 2002). Despite normal ABA levels, and normal responses to other hormones, the null rop10 mutant is hypersensitive to ABA as far as germination is concerned. Freshly harvested rop10-1 seeds germinate at significantly lower rates than wild-type seeds, but after a 4-day moist chilling period, the germination rates of mutant and wild-type seeds are similar. The ABA hypersensitivity of rop10 mutants also extends to root elongation and stomatal closure, and ABA is able to specifically downregulate the synthesis ROP10-encoding transcripts in roots. Constitutive expression of the normal ROP 10 gene in transgenic plants reduces ABA inhibition of seed germination. The negative regulation of ABA signaling by ROP 10 depends on the protein being correctly localized to the plasma membrane, and the protein contains a putative farnesylation site to effect its targeting to this subcellular locale (Zheng and others 2002).
Post-translational Modification and Protein Degradation
Evidence concerning the importance of regulated proteolysis during dormancy breakage is gradually emerging (Bove and others 2005). An example of this also occurs postgerminatively. ABI5, a member of the family of basic leucine zipper transcription factors regulates a subset of LEA genes during seed development and in vegetative tissues in the presence of ABA. It has been implicated in ABA-regulated gene transcription mediated by ABI3/VP1 proteins, and it is known that ABI3 and ABI5 physically interact. ABI5 itself may play a key role during the early postgerminative stages of seedling development; more specifically it re-induces seedling “quiescence” under adverse environmental conditions (Lopez-Molina and others 2001). Under stress conditions, ABI5 accumulates postgerminatively during a 2–3-day period; this depends on the perception of an increase in ABA concentration and is accompanied by a reactivation of embryonic genes. Under these conditions, proteosome-mediated degradation of ABI5 is prevented. An intermediary in this process leading to transcription activation involves mitogen-activated protein kinase signaling (Lu and others 2002). A novel ABI5-interacting protein (AFP, ABI five binding protein) is also induced by ABA, along with ABI5. This binding protein forms high molecular weight complexes with ABI5 in embryo-derived extracts; it is thought to attenuate ABA signals by targeting ABI5 for ubiquitin-mediated degradation in nuclear bodies (Lopez-Molina and others 2003).
DORMANCY TERMINATION IS ACCOMPANIED BY CHANGES IN ABA BIOSYNTHESIS, TURNOVER, AND SENSITIVITY
In general, whether a seed is dormant or quiescent at maturity, both the amount of ABA and the sensitivity of the embryo to ABA will decline during seed development, especially during late maturation and desiccation (Kermode and others 1989; Xu and Bewley 1991; Kermode 1990, 1995; Jiang and others 1996; Bewley 1997; Schmitz and others 2000, 2001, 2002). Genetic studies have clearly demonstrated a function for ABA in dormancy imposition during seed development (see above discussion) (Karssen and others 1983; Kermode 1995; Bewley 1997; Phillips and others 1997; Foley 2001). Moreover, de novo synthesis of ABA appears to be necessary for dormancy maintenance after seed imbibition, for example, in seeds of sunflower (Helianthus annuus), barley, beechnut, lettuce (Lactica sativa), and Douglas-fir (Pseudotsuga menziesii) (Wang and others 1995; Bianco and others 1997; Le Page-Degivry and others 1997; Le Page-Degivry and Garello 1992; Yoshioka and others 1998). For instance, when tobacco seeds are imbibed, there is an accumulation of ABA in dormant seeds but not in seeds that have been allowed to after-ripen. The carotenoid- and ABA-biosynthesis inhibitor fluridone, when used in conjunction with gibberellic acid (GA3), is effective in breaking dormancy; exogenous application of both chemicals to seeds inhibits accumulation of ABA during imbibition (Grappin and others 2000). Similar to tobacco, GA3 and fluridone are effective in breaking dormancy of yellow-cedar seeds in the absence of any additional treatment (Schmitz and others 2001). These seeds normally require a 3-month dormancy-breaking treatment consisting of 1 month of warm, moist conditions followed by 2 months of moist chilling (Ren and Kermode 1999). However, fluridone alone (that is, with no GA3) is much less effective in eliciting germination, indicating that a decline in ABA amount alone is not sufficient to break dormancy, and other changes (for example, synthesis of gibberellins) may also be necessary.
The termination of dormancy (for example, by after-ripening, moist chilling, and smoke) is strongly associated with a decline in ABA content upon transfer to germination conditions, and the decline in ABA corresponds well with a corresponding increase in germination capacity. After-ripening itself has little effect on ABA levels within embryos of dry barley grains (Jacobsen and others 2002). Upon transfer of the after-ripened grain to germination conditions, the ABA content of the embryo declines markedly over the first 12 h; low ABA concentrations are maintained up to 30 h, by which time 90% of the grains have germinated. Dormant grains placed immediately in germination conditions with no previous after-ripening period show some reduction of embryo ABA content over the first 12 h; thereafter, the level increases (for example, between 12 h and 30 h). A similar transient reduction of ABA, followed by its accumulation, occurs in imbibed dormant Arabidopsis seeds of the Cape Verde Islands ecotype (Cvi). This is in marked contrast to imbibed seeds that have received a previous dormancy-breaking treatment (moist-chilling, nitrate, or fluridone), in which ABA levels continue to decline during imbibition (Ali-Rachedi and others 2004). Seeds of the commonly used accessions of Arabidopsis such as Columbia (which are less dormant than seeds of the Cvi ecotype), show a decline in ABA levels upon imbibition in the absence of a previous dormancy-breaking treatment; however, the seeds do not show the large increase in the ABA metabolite ABA glucosylester, which is characteristic of their non-dormant (moist-chilled and imbibed) counterparts (Chiwocha and others 2005).
In Pinus sylvestris (Scots pine) seed, dormancy-breaking treatments that include either white or red light decrease ABA levels prior to radicle protrusion; seeds subjected to a far-red light pulse after red light do not exhibit as great a decline in ABA, nor is dormancy relieved (Tillberg 1992). During dormancy breakage of yellow-cedar seeds, there is about a twofold reduction of ABA in the embryo; in the megagametophyte, ABA does not change; however, the embryos exhibit a change in both ABA turnover and in sensitivity to ABA (Schmitz and others 2000, 2002). Abscisic acid decreases in both the megagametophyte and embryo during moist chilling of Douglas-fir seeds; in the former, ABA declines fourfold during 7 weeks of moist chilling (Corbineau and others 2002). The longer the duration of moist chilling, the faster the rate of ABA decline during subsequent germination (Corbineau and others 2002).
In post-fire environments, smoke can promote the germination of many species that otherwise remain dormant for long periods (Flematti and others 2004; reviewed in Gubler and others 2005). In particular, a butenolide (for example, 3-methyl-2H-furo[2,3-c]pyran-2-one) of smoke is the active stimulator of dormancy-breakage and germination of both fire-dependent and fire-independent species (Flematti and others 2004); its active concentration is less than 10–9 M. The possibility that smoke-related compounds may affect changes in hormone levels has been addressed. For example, dormancy breakage of Nicotiana attenuata seeds induced by smoke-water is accompanied by an eightfold decrease in ABA content of seeds, in marked contrast to seeds subjected to a control (water) treatment for an equivalent period (22 h) (Schwachtje and Baldwin 2004).
Nitric oxide, a gaseous free radical of nitrates is effective in breaking the dormancy of barley and Arabidopsis C24 seeds (Bethke and others 2004). The presence of exogenous ABA interferes with the promotive effects of a nitric oxide donor (sodium nitroprusside) on dormancy termination of seeds of both species, which may indicate that the nitric oxide radical acts upstream of ABA.
Changes in ABA concentration within embryos and surrounding seed tissues can contribute to dormancy inception, maintenance, and termination (Cutler and Krochko 1999; Schmitz and others 2000, 2002). However, it is clear that levels of ABA and ABA catabolites in plant cells and tissues are under constant flux as a result of the opposing forces of biosynthesis versus degradation (reviewed in Cutler and Krochko 1999; Seo and Koshiba 2002). Thus, ABA amount is not likely to be immediately indicative of changes in dormancy status; rather, the capacity for biosynthesis versus catabolism (metabolic flux) is a superior indicator of whether a seed will or will not terminate dormancy (that is, germinate). Increased ABA catabolism is associated with dormancy termination of seeds of yellow-cedar, beechnut, Douglas-fir, and barley (Le Page-Degivry and others 1997; Schmitz and others 2000, 2002; Corbineau and others 2002; Jacobsen and others 2002).
The degradation or inactivation of ABA occurs via oxidation and conjugation (Zaharia and others 2005). The major pathway by which ABA is catabolized is through hydroxylation at the 8′ position to form 8′ hydroxy-ABA, which reversibly cyclizes to phaseic acid (PA). Further reduction of PA can take place at the 4′ position to form dihydrophaseic acid (DPA). Abscisic acid and ABA metabolites (PA and DPA) can also become conjugated with glucose, forming an ester or glucoside. Other minor pathways include formation of 7′ hydroxy-ABA (7′OH-ABA) and ABA 1′,4′ diols (Cutler and Krochko 1999; Zeevaart 1999).
In yellow-cedar seeds, dormancy termination is accompanied by a change in the ability of the embryo to metabolize ABA, in which 8′ hydroxylation becomes rate-limiting (Schmitz and others 2002).
Effective dormancy breakage likely stimulates ABA metabolism, but at the same time may reduce ABA biosynthesis. Moist chilling of Western white pine (Pinus monticola) seeds is accompanied by a significant decrease in ABA in both the embryo and megagametophyte (Figure 2; Feurtado and others 2004). More importantly, the decline of ABA after different durations of moist chilling correlates well with an increased capacity of seeds to germinate after their transfer to germination conditions. The decline of ABA (due to enhanced breakdown and/or reduced synthesis) continues during germination. Thus, moist chilling not only effects a decline in ABA by stimulating ABA metabolism as chilling proceeds but, perhaps equally important, it seems to increase the capacity for ABA catabolism, particularly when the seeds are subsequently placed in germination conditions. Over time, in conditions that are not effective for breaking dormancy, ABA increases in Western white pine seeds, but there is a decrease of PA, 7′OH ABA, and DPA (or maintenance at steady-state amounts), suggesting a slower rate of ABA breakdown. Abscisic acid levels of the embryo and megagametophyte also decrease in response to other effective dormancy-breaking treatments (for example, incubation of mature seeds in the ABA biosynthesis inhibitor fluridone and GA or removal of the seedcoat), and this is accompanied by increases in the metabolites of ABA (Feurtado and others unpublished). It is evident that ABA biosynthesis (that is, a higher capacity for ABA biosynthesis versus catabolism) is important for dormancy maintenance (Figure 3; Feurtado and others 2004) and that the enclosing seed structures influence the embryo’s ability to metabolize ABA in Western white pine seeds (Feurtado and others unpublished).
Changes in ABA and the metabolites PA, DPA, 7′OH ABA, and ABA-GE in embryos of Western white pine seeds during a 98-day moist-chilling treatment, and during and following germination. MC = moist chilling. D1, D3, D6 = days after transfer of seeds to germination conditions. By D6, seeds have germinated and have 0.5–1.0 cm radicles. Based on data from Feurtado and others (2004) with permission from Springer-Verlag.
Model depicting ABA flux and metabolism in Western white pine seeds during dormancy-breakage, germination and growth. As seed populations progress through the different stages, that is, during dormancy termination (moist chilling, 1), the transition to germination (2), and the transition to seedling growth (3), distinct changes in the levels of ABA and ABA metabolites occur. The changes in ABA levels, or any of the metabolites, during moist chilling and germination are the result of shifts in the relative amounts of biosynthesis compared to catabolism. In the Western white pine embryo, ABA is metabolized through both oxidative and conjugative routes; in the megagametophyte, oxidative catabolism is the exclusive metabolic route (Feurtado and others 2004). Transport of ABA and its metabolites can also affect the levels or pools of any given compound. Transport may occur through passive and active processes; routes of transport (for example, between the embryo and the megagametophyte of Western white pine or between seeds and the surrounding medium) are unknown. From Feurtado and others (2004) with permission from Springer-Verlag.
When barley grains are subjected to after-ripening, ABA of the embryo is rapidly metabolized to PA upon subsequent imbibition, and the metabolite does not appear to be released into the incubation medium or into the endosperm (Jacobsen and others 2002). The products of 8’ hydroxylation of ABA (PA and DPA) also accumulate in after-ripened Arabidopsis (Columbia) seeds over the first 24 h of imbibition (Kushiro and others 2004). In previously moist-chilled Arabidopsis (Columbia) seeds, the major product of ABA metabolism is ABA glucose ester; PA and DPA remain at steady state and low levels over the first 48 h of imbibition (Chiwocha and others 2005).
Studies to monitor changes in the amount of ABA in seed tissues should be extended to analyses of the nature of the ABA metabolites. For example, the importance of ABA catabolism via 8′ hydroxylation versus other pathways of catabolism (for example, those involving 7′ or 9′ hydroxylation of ABA) varies between species and even between different tissues and different developmental stages of the same species (Cutler and Krochko 1999; Zeevaart 1999; Zhou and others 2004). This is the case in Western white pine seeds. In the megagametophyte, ABA is metabolized through 8′ hydroxylation, generating PA and DPA; both metabolites increase during a 98-day moist chilling period. In the embryo, ABA is metabolized through both the 8′- and 7′-hydroxylation pathways as well as by conjugation to glucose (Feurtado and others 2004).
Thermodormancy (secondary dormancy) of lettuce seeds, induced by imbibing seeds at a non-optimal temperature for germination (33°C instead of 23°C), is associated with surprisingly active hormone flux. Moreover, the hormone and hormone metabolite profiles of germinating and thermodormant lettuce seeds are distinct. This is particularly true for ABA and its metabolites, in which thermodormant seeds accumulate high levels of DPA, whereas germinating seeds accumulate high amounts of ABA glucose ester. Thermodormant seeds are further distinguished from germinating seeds by exhibiting major accumulations of IAA and zeatin, which are not accompanied by any significant increases in the levels of their conjugates IAAsp and zeatin riboside, respectively. The most striking changes potentially reflective of hormonal cross-talk include a marked accumulation of auxin (IAA) levels in thermodormant seeds that is coincident with a major increase in the level of DPA (Chiwocha and others 2003). Whether there is a key interaction between auxin biosynthesis and ABA catabolism remains to be determined.
Germination of Douglas-fir seeds is negatively affected by oxygen deprivation, and dormant seeds that have not been subjected to a previous moist-chilling treatment are particularly sensitive to oxygen deficiency, especially in comparison to their non-dormant chilled counterparts (Corbineau and others 2002). In isolated embryos of beechnut, the oxidative products of (+)-[3H]-ABA metabolism are reduced as a result of lowering oxygen availability or by incubating embryos in tetcyclasis (a monooxygenase inhibitor). Interestingly, the oxidative products of ABA catabolism are also reduced when embryos are surrounded by the seed coat and pericarp, leading the authors to conclude that these enclosing structures limit oxygen supply to the embryo (Barthe and others 2000). As already noted, the major pathway by which ABA is catabolized —namely hydroxylation at the 8’ position—is accomplished by 8′ hydroxylase, a cytochrome P450 monooxygenase, an enzyme inhibited by oxygen deprivation and tetcyclasis (Krochko and others 1998).
A greater understanding of the relative contributions of ABA biosynthesis and catabolism to dormancy maintenance and its termination will emerge from detailed genetic and biochemical studies of the enzymes that effect dynamic changes in seed and embryo ABA contents during the dormancy-to- germination transition. Most genes involved in ABA biosynthesis have been cloned, although understanding of their regulation and expression in seeds is limited (Nambara and Marion-Poll 2003). Changes in the expression of the zeaxanthin epoxidase gene (ZEP) in Nicotiana plumbaginifolia seeds are generally consistent with the rise and fall in ABA concentration during seed development (Audran and others 1998; reviewed in Seo and Koshiba 2002). The ZEP gene is ubiquitously expressed during the maturation phase of Arabidopsis seed development, but expression becomes restricted to the embryo and endosperm during the desiccation phase (Audran and others 2001). Less information is available for 9-cis epoxycarotenoid dioxygenase (NCED), which exists as a gene family in many species. Several NCED genes are expressed during development of Arabidopsis seeds, and expression persists in the dry seed (Tan and others 2003; Seo and others 2004). The Arabidopsis NCED5 and NCED6 genes are active during seed development until late maturation (Tan and others 2003). This is also true for the Arabidopsis NCED3 gene, the major stress-induced NCED gene; at earlier stages of development, expression is restricted to the basal region of the seed (Tan and others 2003). Abscisic aldehyde oxidase 3 converts ABA aldehyde to ABA (Seo and Koshiba 2002). The gene encoding this enzyme exhibits low expression in Arabidopsis seeds at 10 days after flowering, and transcripts are present in dry seeds (González-Guzmán and others 2004; Seo and others 2004). Detailed expression profiling in relation to dormancy maintenance awaits further investigation. Similarly, the Short Chain Dehydrogenase/Reducase (SDR1), which catalyzes the conversion of xanthoxin to abscisic aldehyde, is expressed at low levels in seeds and embryos (Cheng and others 2002).
The expression of the NCED1 gene is upregulated in embryos during imbibition of both dormant and non-dormant barley grains; however, it is induced to a greater extent in embryos of dormant imbibed grains and may contribute to their higher ABA contents as compared to their non-dormant counterparts. However, another NCED gene (NCED2), by contrast, shows greater expression in embryos of non-dormant imbibed grains (Millar and others unpublished).
For many years the progress toward understanding ABA biosynthesis in seeds has far exceeded that toward understanding the contribution of ABA catabolism to dormancy-breakage and germination, and that differential continues today. However, in recent years, genes encoding enzymes that effect ABA metabolism have been identified. A breakthrough came recently when two groups of investigators identified the cytochrome P450 group, CYP707A, as ABA 8′ hydroxylases in Arabidopsis (Kushiro and others 2004; Saito and others 2004; reviewed in Nambara and Marion-Poll 2005). There are four CYP707A genes in Arabidopsis that exhibit tissue-specific regulation (Kushiro and others 2004; Saito and others 2004). CYP707A2 is the major 8′ hydroxylase gene expressed during seed imbibition; CYP707A2 gene transcripts are present in dry seeds, and expression of this gene peaks at a point approximately 6 h after the start of imbibition in seeds receiving a previous after-ripening period. This timing of expression coincides with the decline in seed ABA and the increase in PA concentration. CYP707A2 gene expression is also induced by 30 μM ABA after 12 h of imbibition (Kushiro and others 2004). In contrast, the expression of the CYP707A1 and A3 genes remains low during early imbibition and increases only after 12 h of imbibition. T-DNA insertional mutants of CYP707A2 have higher seed ABA contents and exhibit increased dormancy as compared to their wild-type counterparts. Thus, CYP707A2 contributes to the reduction of ABA in imbibed after-ripened seeds, a process important for dormancy breakage. However, these analyses were conducted in seeds of the Columbia ecotype, which shows little or no primary dormancy, and it will be important to extend these analyses to a more dormant species. In this respect, the gene encoding a barley 8′ hydroxylase (CYP707A2) is induced to a much higher level in embryos of imbibed non-dormant grains (previously subjected to after-ripening), than in embryos of dormant imbibed grains, and the transcripts (and hence, possibly 8′ hydroxylase action) are localized to the coleorhiza in the region of the primary root tip (Millar and others unpublished).
Two ABA glycosyltransferases have been identified in Arabidopsis and azuki bean (Vigna angularis) (Xu and others 2002; Lim and others 2005). The deduced amino acid sequence of the adzuki bean glucosyltransferase exhibits 30%–44% identity with other known UDP-glucosyltransferase homologs. The gene is ABA-inducible and also increases in response to environmental stress (for example, water stress and wounding). The recombinant enzyme produces primarily ABA-GE from racemic ABA and UDP-glucose.
Despite detailed studies to examine the role of ABA metabolism in dormancy termination and germination, several questions remain to be addressed. One pertains to the distribution of ABA metabolites in different parts of the seed and whether this changes during germination. Does ABA-GE represent an irreversibly inactivated form of ABA, or can it later generate free ABA? It is generally assumed that ABA-GE is not a storage form of ABA and that the conjugation is irreversible, with the conjugate being sequestered in the vacuole (Kleczkowski and Schell 1995; Zeevaart 1999). Although it is clear that ABA is metabolized through various pathways, it is not known what other pathways exist or what the end points of catabolism are. Further, it is not known what the biological roles of the initial catabolites of ABA are (that is, those of 7′OH ABA, 8′OH ABA, and 9′OH ABA) (Zhou and others 2004). It is generally assumed that ABA glucose ester, PA, and DPA are inactive, whereas the hydroxy ABA catabolites (7′ and 8′ hydroxy-ABAs) can act as signaling molecules (Hill and others 1995).
Mechanisms that underlie the fine-tuned balance between ABA biosynthesis and its catabolism and potential transport within and between seed tissues need to be elucidated. Changes in ABA sensitivity (and sensitivity to certain metabolites) undoubtedly also play a role in the transition of seeds from a dormant to a non-dormant state.
References
Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. 2003. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15:2816–2825
Adkins SW, Loewen M, Symons SJ. 1986. Variation within pure lines of wild oats (Avena fatua) in relation to degree of primary dormancy. Weed Sci 34:859–864
Ali-Rachedi S, Bouinot D, Wagner M-H, Bonnet M, Sotta B, et al. 2004. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219:479–488
Amaral da Silva EA, Toorop PE, van Aeist A, Hilhorst HWM. 2004. Abscisic acid controls embryo growth potential and endosperm cap weakening during coffee (Coffea arabica cv. Rubi) seed germination. Planta 220:251–261
Anderberg RJ, Walker-Simmons MK. 1992. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA 89:10183–10187
Audran C, Borel C, Frey A, Sotta B, Meyer C, others. 1998. Expression studies of the zeaxanthin epoxidase gene from Nicotiana plumbaginifolia. Plant Physiol 118:1021–1028
Audran C, Liotenberg S, Gonneau M, North H, Frey A, others. 2001. Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Aust J Plant Physiol 28:1161–1173
Barthe Ph, Garello G, Bianco-Trinchant J, le Page-Degivry MT. 2000. Oxygen availability and ABA metabolism in Fagus sylvatica seeds. J Plant Growth Regul 30:185–191
Bäumlein H, Misera S, Luerben H, Kolle K, Horstmann C, others. 1994. The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6:379–387
Benech-Arnold RL, Fenner M, Edwards PJ. 1991. Changes in the germinability, ABA content and embryonic sensitivity in developing seeds of Sorghum bicolor (L.) Moench. induced by water stress during grain filling. New Phytol 118:339–347
Benech-Arnold RL, Giallorenzi MC, Frank J, Rodriguez V. 1999. Termination of hull-imposed dormancy in barley is correlated with changes in embryonic ABA content and sensitivity. Seed Sci Res 9:39–47
Bethke PC, Gubler F, Jacobsen JV, Jones RL. 2004. Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219:847–855
Bewley JD. 1997. Seed germination and dormancy. Plant Cell 9: 1055–1066
Bewley JD, Black M. 1994. Seeds: physiology of development and germination. 2nd ed. New York, New York, USA, Plenum Publishing Corporation, 445 p
Bianco J, Garello G, Le Page-Degivry MT. 1997. De novo ABA synthesis and expression of seed dormancy in a gymnosperm: Pseudotsuga menziesii. J Plant Growth Regul 21:115–119
Bies-Etheve N, da Silva Conceicao A, Giraudat J, Koornneef M, Léon-Kloosterziel K, et al. 1999. Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol Biol 40:1045–1054
Bove J, Lucas P, Godin B, Ogé L, Jullien M, et al. 2005. Gene expression analysis by cDNA-AFLP highlights a set of new signaling networks and translational control during seed dormancy breaking in Nicotiana plumbaginifolia. Plant Mol Biol 57:593–612
Brocard-Gifford IM, Lynch TJ, Finkelstein RR. 2003. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131:78–92
Calvo AP, Nicolás C, Nicolás G, Rodriguez D. 2004. Evidence of a cross-talk regulation of a GA 20-oxidase (FsGA20ox1) by gibberillins and ethylene during the breaking of dormancy in Fagus silvatica seeds. Physiol Plant 120:623–630
Carrari F, Benech-Arnold R, Osuna-Fernandez R, Hopp E, Sanchez R, et al. 2003. Genetic mapping of the Sorghum bicolor vp1 gene and its relationship with preharvest sprouting. Genome 46:253–258
Carrari F, Perez-Flores L, Lijavetzky D, Enciso S, Sanchez R, et al. 2001. Cloning and expression of a sorghum gene with homology to maize vp1. Its potential involvement in pre-harvest sprouting resistance. Plant Mol Biol 45:631–640
Carson CB, Hattori T, Rosenkrans L, Vasil V, Vasil IK, et al. 1997. The quiescent/colorless alleles of viviparous1 show that the conserved B3 domain of VP1 is not essential for ABA-regulated gene expression in the seed. Plant J 12:1231–1240
Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14:2723–2743
Chiwocha SDS, Abrams SR, Ambrose SJ, Cutler AJ, Loewen S, et al. 2003. A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy in lettuce (Lactuca sativa L.) seeds. Plant J 35:405–417
Chiwocha SDS, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, et al. 2005. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J 42:35–48
Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ. 2002. GCR1, the putative Arabidopsis G protein–coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA 99:4736–4741
Corbineau F, Bianco J, Garello G, Côme D. 2002. Breakage of Pseudotsuga menziesii seed dormancy by cold treatment as related to changes in seed ABA sensitivity and ABA levels. Physiol Plant 114:313–319
Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, et al. 2004. AtGA3ox2, a gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136:3660–3669
Cutler AJ, Krochko JE. 1999. Formation and breakdown of ABA. Trends Plant Sci 4:472–478
Cutler S, Ghassesmian M, Bonetta D, Cooney S, McCourt P. 1996. A protein farnesyl transferase involved in abscisic acid signal transduction. Science 273:1239–1241
Debeaujon I, Koornneef M. 2000. Gibberellin requirement for Arabidopsis seed germination is determined both by testa chacteristics and embryonic abscisic acid. Plant Physiol 122:415–424
Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiol 122:403–414
Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. 2001. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13:853–871
Delseny M, Bies-Etheve N, Carles CH, Hull G, Vicient C, et al. 2001. Late Embryogenesis Abundant (LEA) protein gene regulation during Arabidopsis seed maturation. J Plant Physiol 158:419–427
Derxk MPM, Vermeer E, Karssen C. 1994. Gibberellins in seeds of Arabidopsis thaliana: biological activities, identification and effects of light and chilling on endogenous levels. J Plant Growth Regul 15:223–234
Downie B, Hilhorst HWM, Bewley JD. 1997. Endo-β-mannanase activity during dormancy alleviation and germination of white spruce (Picea glauca) seeds. Physiol Plant 101:405–415
Egerton-Warburton LM. 1998. A smoke-induced alteration of the sub-testa cuticle in seeds of the post-fire recruiter, Emmenanthe penduliflora Benth. (Hydrophyllaceae). J Exp Bot 49:1317–1327
El-Kassaby YA, Chaisurisri K, Edwards DGW, Taylor DW. 1993. Genetic control of germination parameters of Douglas-fir, Sitka spruce, Western red cedar, and yellow-cedar and its impact on container nursery production. In: Edwards DGW, editor. Dormancy and barriers to germination. Proceedings of an International Symposium of IUFRO Project Group P2.04-00 (Seed Problems). Victoria, B.C., Canada: Forestry Canada, Pacific Forestry Centre. p. 37–42
Feurtado JA, Xia J-H, Ma Y, Kermode AR. 2003. Increasing the temperature of the water soak preceding moist chilling promotes dormancy-termination in seeds of Western white pine (Pinus monticola Dougl.). Seed Sci Technol 31:275–288
Feurtado JA, Ambrose SJ, Cutler AJ, Ross ARS, Abrams SR, et al. 2004. Dormancy termination of Western white pine (Pinus monticola Dougl. Ex D. Don) seeds is associated with changes in abscisic acid metabolism. Planta 218:630–639
Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Finkelstein RR, Somerville CR. 1990. Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA response. Plant Physiol 94:1172–1179
Finkelstein RR, Gampala SSL, Rock CD. 2002. Abscisic acid signaling in seeds and seedlings. Plant Cell, Supplement, S15–S45
Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. 2004. A compound from smoke that promotes seed germination. Science 305:977
Foley ME. 2001. Seed dormancy: an update on terminology, physiological genetics, and quantitative trait loci regulating germinability. Weed Sci 49:305–317
Frey A, Audran C, Marin E, Sotta B, Marion-Poll A. 1999. Engineering seed dormancy by modification of zeaxanthin epoxidase gene expression. Plant Mol Biol 39:1267–1274
Frey A, Godin B, Bonnet M, Sotta B, Marion-Poll A. 2004. Maternal synthesis of abscisic acid control seed development and yield in Nicotiana plumbaginafolia. Planta 218:958–964
Fukuhara T, Kirch H-H, Bohnert HJ. 1999. Expression of Vp1 and water channel proteins during seed germination. Plant Cell Environ 22:417–424
Garello G, LePage-Degivry MT. 1999. Evidence for the role of abscisic acid in the genetic and environmental control of dormancy in wheat (Triticum aestivum L.). Seed Sci Res 9:219–226
Giraudat J, Hague BM, Valon C, Smalle J, Parcy F, et al. 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4:1251–1261
Gómez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho T-HD, et al. 1999. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci USA 96:1767–1772
González-Guzmán M, Abia D, Salinas J, Serrano R, Rodríguez PL. 2004. Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiol 135:325–333
Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, et al. 1999. ABI1 protein phosphatase is a negative regulator of abscisic acid signaling. Plant Cell 11:1897–1910
Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M. 2000. Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210:279–285
Groot SPC. 1987. Hormonal regulation of seed development and germination in tomato. Studies on abscisic acid- and gibberellin-deficient mutants. Ph.D. thesis, University of Wageningen, Wageningen, The Netherlands
Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM. 1988. Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seeds prior to radicle protrusion. Planta 174:500–504
Groot SPC, Van Yperen II, Karssen CM. 1991. Strongly reduced levels of endogenous abscisic acid in developing seeds of tomato mutant sitiens do not influence in vivo accumulation of dry matter and storage proteins. Physiol Plant 81:73–78
Grubisic D, Neskowic M, Konjevic R. 1985. Changes in light sensitivity of Paulownia tomentosa and P. fortunei seeds. Plant Sci 39:13–16
Grzelczak ZF, Rahman S, Kennedy TD, Lane BG. 1985. Germin. Compartmentation of the protein, its translable mRNA, and its biosynthesis among roots, stems and leaves of wheat seedlings. Can J Biochem Cell Biol 63:1003–1013
Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV. 2002. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol 129:191–200
Gubler F, Millar AA, Jacobsen JV. 2005. Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8:183–187
Harada JJ. 1997. Seed maturation and control of germination. In: Larkins BA, Vasil IK, (eds.), Advances in cellular and molecular biology of plants, vol. 4, Cellular and molecular biology of seed development. Dordrecht, The Netherlands, Kluwer Academic Publishers, pp. 545–592
Harada JJ. 2001. Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J Plant Physiol 158:405–409
He Y, Gan S. 2004. A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Mol Biol 54:1–9
Hilhorst HWM. 1995. A critical update on seed dormancy. I. Primary dormancy. Seed Sci Res 5:61–73
Hilhorst HWM, Downie B. 1994. Dormancy and germination of abscisic acid-deficient tomato seeds. Further studies with the sitens mutant. Plant Physiol (Supplement) 105:167
Hill A, Nantel A, Rock CD, Quatrano RS. 1996. A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J Biol Chem 271:3366–3374
Hill RD, Liu J-H, Durnin D, Lamb N, Shaw A, Abrams SR. 1995. ABA structure–activity relationships in barley aleurone layers and protoplasts: Biological activity of optically active oxygenated metabolites. Plant Physiol 108:573–579
Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E. 2002. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21:3029–3038
Hoecker U, Vasil IK, McCarty DR. 1995. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev 9:2459–2469
Hoecker U, Vasil IK, McCarty DR. 1999. Signaling from the embryo conditions Vp1-mediated repression of α-amylase genes in the aleurone layer of developing maize seeds. Plant J 19:371–377
Holdsworth M, Kurup S, McKibbin R. 1999. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci 4:275–280
Hugouvieux V, Kwak JM, Schroeder JI. 2001. A mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106:477–487
Hugouvieux V, Murata Y, Young JJ, Kwak JM Mackesy DZ, et al. 2002. Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol 130:1276–1287
Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN. 2002. Abscisic acid, phaseic acid, and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115:428–444
Jacobsen SE, Binkowski KA, Olszewski NE. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93:9292–9296
Jiang L, Abrams S, Kermode AR. 1996. Vicilin and napin storage-protein gene promoters are responsive to abscisic acid in developing transgenic tobacco seed but lose sensitivity following premature desiccation. Plant Physiol 110:1135–1144
Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. 2002. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element- binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130:837–846
Jones HD, Kurup S, Peters NC, Holdsworth MJ. 2000. Identification and analysis of proteins that interact with Avena fatua homologue of the maize transcription factor VIVIPAROUS 1. Plant J 21:133–142
Jones HD, Peters NCB, Holdsworth MJ. 1997. Genotype and environment interact to control dormancy and differential expression of the Viviparous 1 homologue in embryos of Avena fatua. Plant J 12:911–920
Karssen CM. 1995. Hormonal regulation of seed development, dormancy, and germination studied by genetic control. In: Kigel J, Galili G (eds.), Seed development and germination, New York, NY, USA, Marcel Dekker Inc., pp. 333–350
Karssen CM, Lacka E. 1986. A revision of the hormone balance theory of seed dormancy: studies on gibberellin and/or abscisic acid deficient mutants of Arabidopsis thaliana. In: Bopp M (ed.) Plant growth substances, Berlin, Germany, Springer-Verlag, pp. 315–323
Karssen CM, Brinkhorst-Van der Swan DLC, Breekland AD, Koornneef M. 1983. Induction of seed dormancy during seed development by endogenous abscisic acid: studies on abscisic acid-deficient genotypes of Arabidopsis thaliana L. Heynh. Planta 157:158–165
Keith K, Kraml M, Dengler NG, McCourt P. 1994. Fusca 3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6:589–600
Kermode AR. 1990. Regulatory mechanisms involved in the transition from seed development to germination. Crit Rev Plant Sci 9:155–195
Kermode AR. 1995. Regulatory mechanisms in the transition from seed development to germination: interactions between the embryo and the seed environment. In: Kigel J, Galili G (eds.), Seed development and germination, New York, NY, USA, Marcel Dekker Inc., pp. 273–332
Kermode AR, Dumbroff, Bewley JD. 1989. The role of maturation drying in the transition from development to germination VII. Effects of partial and complete desiccation on abscisic acid levels and sensitivity in Ricinus communis L. seeds. J Exp Bot 40:303–313
Kermode AR, Finch-Savage W. 2002. Desiccation sensitivity in orthodox and recalcitrant seeds in relation to development. In: Black M, Pritchard H (eds.), Desiccation and plant survival, Oxford, UK, CABI, pp. 149–184
Kim S, Choi H-I, Ryu H-J, Park JH, Kim MD, et al. 2004. ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol 136:3639–3648
Kirik V, Kölle K, Balzer H-J, Bäumlein H. 1996. Two new oleosin isoforms with altered expression patterns in seeds of the Arabidopsis mutant fus3. Plant Mol Biol 31:413–417
Kirkland K, Johnson E. 1999. Fall seeding: avoiding environmental stress. PBI Bulletin, May, 8–9
Kleczkowski K, Schell J. 1995. Phytohormone conjugates: nature and function. Crit Rev Plant Sci 14:283–298
Koornneef M, Bentsink L, Hilhorst H. 2002. Seed dormancy and germination. Curr Opin Plant Biol 5:33–36
Koornneef M, Reuling G, Karssen CM. 1984. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61:377–383
Krochko JE, Abrams GD, Loewen MK, Abrams SR, Culter AJ. 1998. (+)-Abscisic acid 8’ hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol 118:849–860
Kroj T, Savino G, Valon C, Giraudat J, Parcy F. 2003. Regulation of storage protein gene expression in Arabidopsis. Development 130:6065–6073
Kuhn JM, Schroeder JI. 2003. Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6:463–469
Kurup S, Jones HD, Holdsworth MJ. 2000. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21:143–155
Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, et al. 2004. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: key enzymes in ABA catabolism. EMBO J 23:1647–1656.
Kwak JM, Moon J-H, Murata Y, Kuchitsu K, Leonhardt N, et al. 2002. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14:2849–2861
Lazarova G, Zeng Y, Kermode AR. 2002. Cloning and expression of an ABSCISIC ACID INSENSITIVE 3 (ABI3) gene homologue of yellow-cedar (Chamaecyparis nootkatensis). J. Exp Bot 53:1219–1221
Le Page-Degivry MT, Garello G. 1992. In situ abscisic acid synthesis. A requirement for induction of embryo dormancy in Helianthus annus. Plant Physiol 98:1386–1390
Le Page-Degivry MT, Garello G, Barthe P. 1997. Changes in abscisic acid biosynthesis and catabolism during dormancy breaking in Fagus sylvatica embryo. J Plant Growth Regul 16:57–61
Leadem C. 1990. The role of plant growth regulators in the germination of forest tree seeds. J Plant Growth Regul 6:61–93
Lee S, Cheng H, King KE, Wang W, He Y, others. 2002. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16:646–658
Léon-Kloosterziel KM, Van de Bunt GA, Zeevaart JAD, Koornneef M. 1996. Arabidopsis mutants with reduced seed dormancy. Plant Physiol 110:233–240
Leube MP, Grill E, Amrhein N. 1998. ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett 424:100–104
Leubner-Metzger G, Meins F Jr. 2000. Sense transformation reveals a novel role for class I β-1,3-glucanase in tobacco seed germination. Plant J 23:215–221
Leubner-Metzger G, Fründt C, Vögeli-Lange R, Meins F Jr. 1995. Class I β-1,3-glucanases in the endosperm of tobacco during germination. Plant Physiol 109:751–759
Leung J, Bouvier-Durnd M, Morris PC, Guerrier D, Chefdor F, et al. 1994. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264:1448–1452
Leung J, Merlot S, Giraudat J. 1997. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9:759–771
Leurssen H, Kirik V, Herrmann P, Misera S. 1998. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15:755–764
Li G, Chandrasekharan MB, Wolffe AP, Hall TC. 2001. Chromatin structure and phaseolin gene regulation. Plant Mol Biol 46:121–129
Lim E-K, Doucet CJ, Hou B, Jackson RG, Abrams SR, et al. 2005. Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron Asymmetr 16:143–147
Lopez-Molina L, Mongrand S, Chua N-H. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98:4782–4787
Lopez-Molina L, Mongrand S, Kinoshita N, Chua N-H. 2003. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17:410–418
Lorenzo O, Rodríguez D, Nicolás G, Rodríguez PL, Nicolás C. 2001. A new protein phosphatase 2C (FsPP2c1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiol 125:1949–1956
Lotan T, Ohto M, Yee KM, West MAL, Lo R, et al. 1998. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205
Lu C, Han M-H, Guevara-Garcia A, Fedoroff NV. 2002. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 99:15812–15817
McCarty DR. 1995. Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol 46:71–93
McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, et al. 1991. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Plant Cell 66:895–905
McCourt P, 2001. Plant hormone signaling: getting the message out. Molecules Cells 8:1157–1158
McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, et al. 2003. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15:1120–1130
McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, et al. 2002. Transcripts of Vp-1 homologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci USA 99:10203–10208
Meinke DW. 1995. Molecular genetics of plant embryogenesis. Annu Rev Plant Physiol Plant Mol Biol 46:369–394
Meinke DW, Franzmann LH, Nickle TC, Yeung EC. 1994. Leafy cotyledon mutants of Arabidopsis. Plant Cell 6:1049–1064
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. 2001. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25:295–303
Meyer K, Leube MP, Grill E. 1994. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264:1452–1455
Nakamura S, Toyama T. 2001. Isolation of a VP1 homologue from wheat and analysis of its expression in embryos of dormant and non-dormant cultivars. J Exp Bot 52:875–876
Nakamura S, Lynch TJ, Finkelstein RR. 2001. Physical interactions between ABA response loci of Arabidopsis. Plant J 26:627–635
Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, others. 2000. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 220:412–423
Nambara E, Keith K, McCourt P, Naito S. 1995. A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121:629–636
Nambara E, Marion-Poll A. 2003. ABA action and interactions in seeds. Trends Plant Sci 8:213–217
Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Ogas J, Cheng J-C, Sung ZR, Somerville C. 1997. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277:91–94
Ogawa M, Hanada A, Yamauchi, Kuwahara Y, Kamiya Y, others. 2003. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15:1591–1604
Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. 1993. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana—a comparative study using abscisic acid insensitive abi3 mutants. Plant Physiol 102:1185–1191
Osa M, Kato K, Mori M, Shindo C, Torada A, et al. 2003. Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet 106:1491–1496
Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, et al. 2005. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17:1105–1119
Paek NC, Lee B-M, Bai DG, Smith JD. 1998. Inhibition of germination gene expression by viviparous-1 and ABA during maize kernel development. Molecules Cells 8:336–342
Papi M, Sabatini S, Altamura MM, Hennig L, Schäfer E, et al. (2002) Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol 128:411–417
Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, et al. 2000. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev 14:28–33
Parcy F, Valon C, Kohara A, Misera S, Giraudat J. 1997. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert in controlling multiple aspects of Arabidopsis seed development. Plant Cell 9:1265–1277
Peng J, Carol P, Richards DE, Ling KE, Cowling RJ, et al. 1997. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205
Pérez-Flores L, Carrari F, Osuna-Fernández R, Rodriguez MV, Enciso S, et al. 2003. Expression analysis of a GA 20-oxidase in embryos from two sorghum lines with contrasting dormancy: possible participation of this gene in the hormonal control of germination. J Exp Bot 54:2071–2079
Phillips J, Artsaenko O, Fiedler U, Horstmann C, Mock HP, et al. 1997. Seed-specific immunomodulation of abscisic acid activity induces a developmental switch. EMBO J 16:4489–4496
Razem FA, Luo J-H, Abrams SR, Hill RD. 2004. Purification and characterization of a barley aleurone abscisic acid-binding protein. J Biol Chem 279:9922–9929
Reidt W, Wohlfarth T, Elleström M, Czihal A, Tewes A, et al. 2000. Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21:401–408
Ren C, Kermode AR. 1999. Analyses to determine the role of the megagametophyte and other seed tissues in dormancy maintenance of yellow cedar (Chamaecyparis nootkatensis) seeds: morphological, cellular and physiological changes following moist chilling and during germination. J Exp Bot 50:1403–1419
Ren C, Kermode AR. 2000. An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol 124:231–242
Richards DE, King KE, Ait-ali T, Harberd NP. 2001. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52:67–88
Rodriguez PL, Leube MP, Grill E. 1998. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol 38:879–883
Romagosa I, Prada D, Moralejo MA, Sopena A, Muñoz P, et al. 2001. Dormancy, ABA content and sensitivity of a barley mutant to ABA application during seed development and after ripening. J Exp Bot 52:1499–1506
Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, et al. 2004. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydoxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134:1439–1449
Schmitz N, Abrams SR, Kermode AR. 2000. Changes in abscisic acid content and embryo sensitivity to (+)-abscisic acid during dormancy termination of yellow-cedar seeds. J Exp Bot 51:1159–1162
Schmitz N, Abrams SR, Kermode AR. 2002. Changes in ABA turnover and sensitivity that accompany dormancy termination of yellow-cedar (Chamaecyparis nootkatensis) seeds. J Exp Bot 53:89–101
Schmitz N, Xia J-H, Kermode AR. 2001. Dormancy of yellow-cedar seeds is terminated by gibberellic acid in combination with fluridone or with osmotic priming and moist chilling. Seed Sci Technol 29:331–346
Schultz TF, Medina J, Hill A, Quatrano RS. 1998. 14-3-3 proteins are part of an abscisic acid-VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell 10:837–847
Schwachtje J, Baldwin IT. 2004. Smoke exposure alters endogenous gibberellin and abscisic acid pools and gibberellin sensitivity while eliciting germination in the post-fire annual, Nicotiana attenuata seed. Seed Sci Res 14:51–60
Seo M, Koshiba T. 2002. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7:41–48
Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, et al. 2004. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45:1694–1703
Sheen J. 1998. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95:975–980
Silverstone AL, Ciampaglio CN, Sun T-P. 1998. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10:155–169
Söderman E, Hjellstrom M, Fahleson J, Engstrom P. 1999. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol Biol 40:1073–1083
Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR. 2000. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124:1752–1765
Steber C, Cooney SE, McCourt P. 1998. Isolation of the GA-response mutant sly1 as a suppressor of ABI1 in Arabidopsis thaliana. Genetics 149:509–521
Steinbach HS, Benech-Arnold R, Kristof G, Sánchez RA, Marcucci-Poltri S. 1995. Physiological basis of pre-harvest sprouting resistance in Sorghum bicolor (L.) Moench. ABA levels and sensitivity in developing embryos of sprouting-resistant and -susceptible varieties. J Exp Bot 46:701–709
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, et al. 2001. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98:11806–11811
Sussex IM. 1975. Growth and metabolism of the embryo and attached seedlings of the viviparous mangrove, Rhizophora mangle. Am J Bot 62:948–953
Suzuki M, Kao CY, Cocciolone S, McCarty DR. 2001. Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28:409–418
Tahtiharju S, Palva T. 2001. Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26:461–470
Tan B-C, Joseph LM, Deng W-T, Liu L, et al. 2003. Molecular characterization of the Arabidopsis 9-cis-epoxycarotenoid dioxygenase gene family. Plant J 35:44–56
Tan BC, Schwartz SH, Zeevaart JAD, McCarty D. 1997. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94:12235–12240
Taylor IB, Sonneveld T, Bugg TDH, Thompson AJ. 2005. Regulation and manipulation of the biosynthesis of abscisic acid, including the supply of xanthophyll precursors. J Plant Growth Regul 24:253–273
Terskikh VV, Feurtado JA, Ren C, Abrams SR, Kermode AR. 2005. Water uptake and oil distribution during imbibition of seeds of Western white pine (Pinus monticola Dougl. Ex D. Don) monitored in vivo using magnetic resonance imaging. Planta 221:17–27
The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of flowering plant Arabidopsis thaliana. Nature 408:796–815
Tillberg E. 1992. Effect of light on abscisic acid content in photosensitive Scots pine (Pinus sylvestris L.) seed. J Plant Growth Regul 11:147–152
Tyler L., Thomas SG, HU J, Dill A, Alonso JM, et al. 2004. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135:1008–1019
Walker-Simmons M. 1990. Dormancy in cereals—levels of and response to abscisic acid. In: Pharis RP, Rood SB (eds.), Plant growth substances. New York, NY USA, Springer-Verlag, pp. 400–406
Walker-Simmons MK. 1987. ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol 84:61–66
Wang M, Heimovaara-Dijkstra S, Van Duijn B. 1995. Modulation of germination of embryos isolated from dormant and non-dormant barley grains by manipulation of endogenous abscisic acid. Planta 195:586–592
White CN, Riven CJ. 2000. Gibberellins and seed development in maize. II. Gibberellin synthesis inhibition enhances abscisic acid signaling in cultured embryos. Plant Physiol 122:1089–1097
White CN, Proebsting WM, Hedden P, Rivin CJ. 2000. Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol 122:1081–1088
Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, et al. 2001. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1:771–781
Xu N, Bewley JD. 1991. Sensitivity to abscisic acid and osmoticum changes during embryogenesis of alfalfa (Medicago sativa). J Exp Bot 42:821–826
Xu Z-J, Nakajima M, Suzuki Y, Yamaguchi I. 2002. Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from Adzuki bean seedlings. Plant Physiol 129:1285–1295
Yang Z. 2002. Small GTPases: versatile signaling switches in plants. Plant Cell (Supplement) 14:S375–S388
Yoshioka T, Endo T, Satoh S. 1998. Restoration of seed germination at supraoptimal temperature by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol 39:307–312
Zaharia LI, Walker-Simmon MK, Rodríguez CN, Abram SR. 2005. Chemistry of abscisic acid and abscisic acid catabolites and analogs. J Plant Growth Regul XX:xxx–xxx
Zeevaart JAD. 1999. Abscisic acid metabolism and its regulation. In: Hooykaas PJJ, Hall KR, Libbenga KR (eds.), Biochemistry and molecular biology of plant hormones. Amsterdam, The Netherlands, Elsevier Science BV, pp. 189–207
Zeng Y, Raimondi N, Kermode AR. 2003. Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant Mol Biol 51:39–49
Zeng Y, Kermode AR. 2004. A gymnosperm ABI3 gene functions in a severe abscisic acid-insensitive mutant of Arabidopsis (abi3-6) to restore the wild-type phenotype and demonstrates a strong synergistic effect with sugar in the inhibition of post-germinative growth. Plant Mol Biol 56:731–746
Zentella R, Yamauchi D, Ho T-HD. 2002. Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14:2289–2301
Zhang DP, Wu ZY, Li XY, Zhao ZX. 2002. Purification and identification of a 42- kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol 128:714–725
Zheng Z-L, Nafisi M, Tam A, Li H, Crowell DN, et al. 2002. Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14:2787–2797
Zhou R, Cutler AJ, Ambrose SJ, Galka MM, Nelson KM, et al. 2004. A new abscisic acid catabolic pathway. Plant Physiol 134:361–369
Zou X, Seemann JR, Neuman D, Shen QJ. 2004. A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J Biol Chem 279:55770–55779
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kermode, A. Role of Abscisic Acid in Seed Dormancy. J Plant Growth Regul 24, 319–344 (2005). https://doi.org/10.1007/s00344-005-0110-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-005-0110-2