Abstract
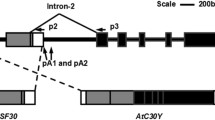
Exposure of cold-hardy Rubidoux trifoliate orange [Poncirus trifoliata (L) Raf.] plants to temperatures from 28°C to −5°C enabled us to isolate and characterize a novel citrus low-temperature gene (CLT) with two transcripts, called CLTa and CLTb, from leaves and stems. CLTa was produced when plants were subjected to low temperatures (starting at 10°C), while CLTb was constitutively expressed. Both CLTa and CLTb have the same open reading frame (ORF) of 165 nucleotides and encode a small (54 deduced amino acid) protein. However, CLTa has an additional 98 nucleotides in the 3′-untranslated region (UTR) that are absent in CLTb. Expression analysis using relative quantitative RT-PCR demonstrated that CLTa is expressed exclusively at low temperatures, while CLTb is expressed constitutively (expression verified from 33°C to −5°C). A GenBank database search identified 61 nucleotides inside of the ORF that are highly similar to low-temperature-responsive genes from Arabidopsis thaliana and Solanum tuberosum. The deduced amino acid sequence revealed similarity with low-temperature-responsive proteins from A. thaliana, Oryza sativa, and S. tuberosum of 77%, 81%, and 73.9%, respectively. A genomic clone was isolated, and the genome organization revealed the presence of three exons and two introns, the second of which is in the 3′ UTR and participates in alternative 3′ splice site selection. One of the 3′ splice sites of the second intron was located immediately before the additional 98-bp non-coding fragment of CLTa, and the second at the very end of the 98-bp fragment. Additionally, the presence of the tetranucleotides TCTT and TTCT, which are involved in the regulation of transcript processing in animals and possibly also active in peach, was found in this intron. Competition for splicing sites on the pre-mRNA in the spliceosome, which is induced by low temperature, may be involved in the production of the two transcripts of the CLT gene.





Similar content being viewed by others
Abbreviations
- cDNA :
-
Complementary DNA
- CLT :
-
Citrus low temperature
- LTR :
-
Low temperature responsive
- ORF :
-
Open reading frame
- UTR :
-
Untranslated region
References
Ayliffe MA, Frost DV, Finnegan EJ, Lawrence GJ, Anderson PA, Ellis JG (1999) Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J 17:287–292
Bassett C, Artlip TS, Callahan AM (2002) Characterization of the peach homologue of the ethylene receptor, PpETR1, reveals some unusual features regarding transcript processing. Planta 215:679–688
Bournay AS, Hedley PE, Maddison A, Waugh R, Machray GC (1996) Exon skipping induced by cold stress in a potato invertase gene transcript. Nucleic Acids Res 24:2347–2351
Brooks SA, Rigby WFC (2000) Characterization of the mRNA ligands bound by the RNA binding protein hnRNP A2 utilizing a novel in vivo technique. Nucleic Acids Res 28:e49
Brown JWS, Simpson CG (1998) Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol 49:77–95
Cai Q, Moore GA, Guy CL (1995) An unusual group of 2 LEA gene family in citrus responsive to low temperature. Plant Mol Biol 29:11–23
Cattivelli L, Bartels D (1990) Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiol 9:1504–1510
Chen CY, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20:465–470
Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995
Côté J, Dupuis S, Wu JY (2001) Polypyrimidine tract-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J Biol Chem 276:8535–8543
Dearth LR, DeWille J (2003) An AU-rich element in the 3′ untranslated region of the C/EBPδ mRNA is important for protein binding during G0 growth arrest. Biochem Biophys Res Commun 304:344–350
Gilmour SJ, Arus NN, Thomashow MF (1992) cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol Biol 18:13–21
Guy CL (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41:187–223
Hara M, Wakasugi Y, Ikoma Y, Yono M, Ogawa K, Kuboi T (1999) cDNA sequence and expression of a cold-responsive gene in Citrus unshiu. Biosci Biotechnol Biochem 63:433–437
Hughes MA, Dunn MA (1996) The molecular biology of plant acclimation to low temperature. J Exp Bot 47:291–305
Karcher D, Bock R (2002) Temperature sensitivity of RNA editing and intron splicing reactions in the plastid ndhB transcript. Curr Genet 41:48–52
Kirch HH, vanBerkel J, Glaczinski H, Salamini F, Gebhardt CH (1997) Structural organization, expression and promoter activity of a cold-stress-inducible gene of potato (Solanum tuberosum L.). Plant Mol Biol 33:897–909
Kurihara-Yonemoto S, Handa H (2001) Low temperature affects the processing pattern and RNA editing status of the mitochondrial cox2 transcripts in wheat. Curr Genet 40:203–208
Kurkela S, Borg-Franck M (1992) Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol 19:689–692
Kurkela S, Franck M (1990) Cloning and characterization of a cold- and ABA-induced Arabidopsis gene. Plant Mol Biol 15:137–144
Le Guiner C, Plet A, Galiana D, Gesnel MC, Del Gatto-Konczak F, Breathnach R (2001) Polypyrimidine tract-binding protein represses splicing of a fibroblast growth factor receptor-2 gene alternative exon through exon sequences. J Biol Chem 23:43677–43687
Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967–971
Lorković ZJ, Kirk DAW, Lambermon MHL, Filipowicz W (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci 5:160–167
Macknight R, Bancroft L, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Mano S, Hayashi M, Nishimura M (1999) Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J 17:309–320
Nambiar MP, Enyedy EJ, Warke VG, Krishnan S, Dennys G, Kammer GM, Tsokos GC (2001) Polymorphism/mutations of the TCR-ζ-chain promoter and 3′ untranslated region and selective expression of TCR-ζ-chain with an alternative spliced 3′ untranslated region in patients with systemic lupus erythematosus. J Autoimmunol 16:133–142
Nordin K, Vahala T, Palva ET (1993) Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 21:641–653
Pawlowski K, Twigg P, Dobritsa S, Guan C, Mullin BC (1997) A nodule-specific gene family from Alnus glutinosa encodes glycine- and histidine- rich proteins expressed in the early stages of actinorhizal nodule development. Mol Plant Microbe Interact 10:656–664
Pearce RS (1999) Molecular analysis of acclimation to cold. Plant Growth Regul 29:47–76
Peng H, Lever E (1995) Regulation of Na+ -coupled glucose transport in LLC-PK1 cells. J Biol Chem 270:23996–24003
Peynado A, Cooper WC (1963) A comparison of three major Texas freezes and tissue temperatures of citrus tree parts during the January 9 to 12 freeze. Proc Rio Grande Valley Hortic Soc 17:15–23
Rouse RE (1985) Review of citrus rootstock for Texas following the 1983 freeze. J Rio Grande Valley Hortic Soc 38:19–26
Rousseau P, Le Discord M, Mouillot G, Marcou C, Carosella ED, Moreau P (2003) The 14 bp deletion-insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol 64:1005–1010
Schaaf MJM, Cidlowski JA (2002) AUUUA motifs in the 3′ UTR of human glucocorticoid receptor α and β mRNA destabilize mRNA and decrease receptor protein expression. Steroids 67:627–636
Smith CWJ, Patton JG, Nadal-Ginard B (1989) Alternative splicing in the control of gene expression. Annu Rev Genet 23:527–577
Southby J, Gooding C, Smith CWJ (1999) Polypyrimidine tract-binding protein functions as a repressor to regulate alternative splicing of α-actinin mutually exclusive exons. Mol Cell Biol 19:2699–2711
Thomashow MF (1990) Molecular genetics of cold acclimation in high plants. Adv Genet 28:99–131
Tignor ME, Davies FS, Sherman WB (1997) Rapid freeze acclimation of Poncirus trifoliata seedlings exposed to 10°C and long days. HortScience 32:854–857
Tignor ME, Davies FS, Sherman WB (1998) Freezing tolerance and growth characteristics of USDA intergeneric citrus hybrids US 119 and selection 17-11. HortScience 33:744–748
Vu JCV, Gupta SK, Yelenosky G, Ku MSB (1995) Cold induced changes in ribulose 1,5-bisphosphate carboxylase-oxygenase and phosphoenolpyruvate carboxylase in citrus. Environ Exp Bot 35:25–31
Weiser CJ (1970) Cold resistance and injury in woody plants. Science 169:1269–1278
Yelenosky G, Hearn CJ, Cooper WC (1968) Relative growth of trifoliate orange selections. Proc Fla State Hortic Soc 93:205–209
You Y, Chen CY, Shyu AB (1992) U-rich sequence-binding proteins (URBPs) interacting with a 20 nucleotide U-rich sequence in the 3′ untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol 12:2931–2940
Zhang Z, Honda C Kita M, Nakayma M, Moriguchi T (2003) Structure and expression of spermidine synthase genes in apple: two cDNAs are spatially and developmentally regulated through alternative splicing. Mol Genet Genomics 268:799–807
Acknowledgements
This research was funded in part by the Texas Citrus Producers Board and by USDA Multicultural Alliances Hispanic-Serving Institutions Education Grants.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Ozias-Akins
Rights and permissions
About this article
Cite this article
Jia, Y., del Rio, H.S., Robbins, A.L. et al. Cloning and sequence analysis of a low temperature-induced gene from trifoliate orange with unusual pre-mRNA processing. Plant Cell Rep 23, 159–166 (2004). https://doi.org/10.1007/s00299-004-0805-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-004-0805-z