Summary
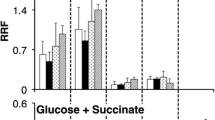
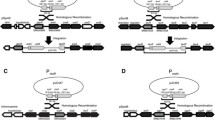
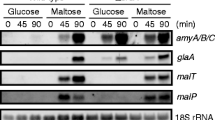
Both the MAL1 and MAL6 loci in Saccharomyces strains have been shown by functional and structural studies to comprise a cluster of at least three genes necessary for maltose utilization. They include regulatory, maltose transport and maltase genes designated MALR, MALT and MALS, respectively. Subclones of each gene derived from the MAL6 locus were inserted into the multicopy shuttle plasmid YEp13, introduced into MAL1 and mal1 strains and the effects of altered gene dosage of each gene, or a combination of them, on MAL gene expression investigated. MAL1 strains transformed with a plasmid carrying the MAL6S gene showed coordinate four to five fold increases in both maltase enzyme activity and its mRNA, whereas no increase in maltose transport activity or of MALT mRNA was observed when MAL6T was present on multicopy plasmids. The presence of the MAL6R gene on a multicopy plasmid led to greatly increased transcription of both inducible and constitutive mRNAs with homology to the regulatory gene; it also gave rise to two fold increases in both induced maltase mRNA levels and enzyme activity, but only in the presence of maltose. However, it had no apparent effect on the accumulation of MALT mRNA. Finally, the induction kinetics of plasmid-borne and chromosomal MALS and MALT gene expression were examined under conditions of altered gene dosage of the MAL6 regulatory and structural genes. The results of these experiments indicate that MALR encodes a trans-acting positive activator that requires maltose for induction of MALS and MALT transcription even when the regulatory gene is present on a multicopy plasmid. Maltose transport can be a rate-limiting factor in MAL gene expression, at least in the early stages of induction. The regulation of the MALS and MALT genes, whose activities are coordinately induced in MAL1 strains by maltose, may in fact exhibit some important differences.
Similar content being viewed by others
References
Baker SM, Okkema PG, Jaehning JA (1984) Expression of the Saccharomyces cerevisiae GAL7 gene on autonomous plasmids. Mol Cell Biol 4:2062–2071
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Brent R, Ptashne M (1985) A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43:729–736
Chevallier MR (1982) Cloning and transcriptional control of a eukaryotic permease gene. Mol Cell Bil 2:977–984
Chow T (1987) Structure and organization of the yeast MAL loci. Doctoral Dissertation, Albert Einstein College of Medicine, Yeshiva University
Chow T, Goldenthal MJ, Cohen JD, Hegde M, Marmur J (1983) Identification and physical characterization of yeast maltase structural genes. Mol Gen Genet 191:366–371
Cohen JD, Goldenthal MJ, Buchferer B, Marmur J (1984) Mutational analysis of the MAL1 locus of Saccharomyces: identification and functional characterization of three genes. Mol Gen Genet 196:208–216
Cohen JD, Goldenthal MJ, Chow T, Buchferer B, Marmur J (1985) Organization of the MAL loci of Saccharomyces. Physical identification and functional characterization of three genes at the MAL6 locus. Mol Gen Genet 200:1–8
Cove DJ (1969) Evidence for a near limiting intracellular concentration of a regulator substance. Nature 224:272–273
deKroon RA, Koningsberger W (1970) An inducible transport system for α-glucosides in protoplasts of Saccharomyces carlsbergensis. Biochim Biophys Acta 204:590–609
delaFuente G, Sols A (1962) Transport of sugars in yeast II. Mechanisms of utilization of disaccharides and related glycosides. Biochim Biophys Acta 56:49–62
Dubin RA, Perkins EL, Needleman RB, Michels CA (1986) Identification of a second trans-acting gene controlling maltose fermentation in Saccharomyces carlsbergensis. Mol Cell Biol 6:2757–2765
Elion EA, Warner JR (1984) The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39:663–673
Federoff JH, Eccleshall TR, Marmur J (1983a) Regulation of maltase synthesis in Saccharomyces carlsbergensis. J Bacteriol 154:1301–1308
Federoff HJ, Eccleshall TR, Marmur J (1983b) Carbon catabolite repression of maltase synthesis in Saccharomyces carlsbergensis. J Bacteriol 156:301–307
Goldenthal MJ, Cohen JD, Marmur J (1983) Isolation and characterization of a maltose transport mutant in Saccharomyces cerevisiae. Curr Genet 7:195–199
Guarente L (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol 101:181–191
Guarente L (1984) Yeast promoters: positive and negative elements. Cell 36:799–800
Guerry P, LeBlanc DJ, Falkow S (1973) General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol 116:1064–1066
Hashimoto H, Kikuchi Y, Nogi Y, Fukasawa T (1983) Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae: Isolation and characterization of the regulatory gene GAL4. Mol Gen Genet 191:31–38
Hong SH, Marmur J (1986) Primary structure of the maltase gene of the MAL6 locus of Saccharomyces carlsbergensis. Gene 41:75–84
Ito H, Fukada Y, Murata K, Kimura K (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168
Johnston SA, Hopper JE (1982) Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effect on the galactose melibiose regulon. Proc Natl Acad Sci USA 79:6971–6975
Keegan L, Gill G, Ptashne M (1986) Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231:699–704
Koren R, LeVitre JA, Bostian KA (1986) Isolation of the positive-acting regulatory gene PHO4 from Saccharomyces cerevisiae. Gene 41:271–280
Laughon A, Gesteland RF (1982) Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc Natl Acad Sci USA 79:6827–6831
Losson R, Fuchs RPP, Lacroute F (1983) In vivo transcription of a eukaryotic regulatory gene. EMBO J 2:2179–2184
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Marczynski GT, Jaehning JA (1985) A transcription map of a yeast centromere plasmid: unexpected transcripts and altered gene expression. Nucleic Acids Res 13:8487–8506
Michels CA, Needleman RB (1984) The dispersed, repeated family of MAL loci in Saccharomyces spp. J Bacteriol 157:949–952
Mowshowitz DB (1979) Gene dosage effects on the synthesis of maltase in yeast. J Bacteriol 137:1200–1207
Mueller PP, Hinnebusch AG (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell 45:201–207
Murray NE, Bruce SA, Murray K (1979) Molecular cloning of the DNA ligase gene from bacteriophase T4. II. Amplification and preparation of the gene product. J Mol Biol 132:493–505
Needleman RB, Kaback DB, Dubin RA, Perkins EL, Rosenberg NG, Sutherland KA, Forrest DB, Michels CA (1984) MAL6 of Saccharomyces: a complex locus containing three genes required for maltose fermentation. Proc Natl Acad Sci USA 81:2811–2815
Ng R, Abelson J (1980) Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 77:3912–3916
Parent SA, Fenimore CM, Bostian KA (1985) Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast 1:83–138
Raibaud O, Schwartz M (1984) Positive control of transcription initiation in bacteria. Annu Rev Genet 18:173–206
Savageau MA (1979) Autogenous and classical regulation of gene expression. A general theory and experimental evidence. In: Goldberger RF (ed) Biological regulation and development, vol 1. Gene expression. Plenum, New York
Serrano R (1977) Energy requirements for maltose transport in yeast. Eur J Biochem 80:97–102
Sherman F, Fink GR, Lawrence CW (1972) Laboratory manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Tschopp JF, Emr S, Field C, Schekman R (1986) GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J Bacteriol 166:313–318
Warner JR, Mitra G, Schwindinger WF, Studeny M, Fried H (1985) Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol 5:1512–1521
Author information
Authors and Affiliations
Additional information
Communicated by C.P. Hollenberg
Rights and permissions
About this article
Cite this article
Goldenthal, M.J., Vanoni, M., Buchferer, B. et al. Regulation of MAL gene expression in yeast: Gene dosage effects. Mol Gen Genet 209, 508–517 (1987). https://doi.org/10.1007/BF00331157
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00331157