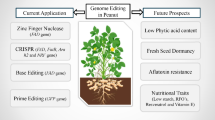
Abstract
Pearl millet [Cenchrus americanus (L.) Morrone; also known as Pennisetum glaucum], originated 4900 years ago, is a C4 crop with high photosynthetic efficiency and fulfills the food and fodder needs of resource-poor farmers of sub-Saharan Africa, Southeast Asia, and the Indian subcontinent. Pearl millet is a climate-ready crop and grows well in poor and low-fertility soil. It is profoundly nutritious, fiber-rich, and non-glutinous. Based on transcriptome and bioinformatics studies, it is estimated that 1.79 Gb of the pearl millet genome consists of 38,579 genes. Unfortunately, functional genomics and genotype-phenotype association in pearl millet are poorly explored areas. Pearl millet suffers from low yield for many reasons. There is an urgent need to validate the functions of important genes to improve the crop and better utilize it for future agriculture in a changing climate scenario. In recent years, genome editing, especially CRISPR-Cas, has come under the spotlight for improving crop varieties. In this chapter, we discuss how the available genome editing tools can play a significant role in deciphering the functions of pearl millet genomic regions and crop improvement. We also highlight major bottlenecks to using genome editing in pearl millet and discuss possible ways to overcome those constraints.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- ABEs:
-
Adenine base editors
- CBEs:
-
Cytosine base editors
- CGBEs:
-
C to G base editors
- CRISPR-Cas:
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein
- DSBs:
-
Double-strand breaks
- HDR:
-
Homology-directed repair
- MMEJ:
-
Microhomology mediated end joining
- NHEJ:
-
Non-homologous end joining
- pegRNA:
-
Prime editing gRNA
- RFLP:
-
Restriction fragment length polymorphism
- SNPs:
-
Single nucleotide polymorphisms
- SSN:
-
Sequence-specific nuclease
- TALENs:
-
Transcription activator like effector nucleases
- ZFNs:
-
Zinc finger nucleases
References
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM et al (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576(7785):149–157
Burgarella C et al (2018) A Western Sahara Centre of domestication inferred from pearl millet genomes. Nat Ecol Evol 2:1377–1380
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Choudhary M, Jayanand, Padaria JC (2015) Transcriptional profiling in pearl millet (Pennisetum glaucum L.R. Br.) for identification of differentially expressed drought responsive genes. Physiol Mol Biol Plants 21:187–196
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F et al (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genet 186:757–761
Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF et al (2020) A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol 38:1274–1279
Debieu M, Kanfany G, Laplaze L (2017) Pearl millet genome: lessons from a tough crop. Trends Plant Sci 22(11):911–913
Diack O, Kanfany G, Gueye MC, Sy O, Fofana A, Tall H et al (2020) GWAS unveils features between early-and late-flowering pearl millets. BMC Genomics 21(1):1–11
Dong H et al (2020) Generation of imidazolinone herbicide resistant trait in Arabidopsis. PLoS One 15:e0233503
Dwivedi S, Upadhyaya H, Senthilvel S, Hash C (2012) Millets: genetic, and genomic resources. In: Janick J (ed) Plant breeding reviews. Wiley, Hoboken, pp 247–375
FAO (2017) The future of food and agriculture: trends and challenges. FAO
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23(10):1229–1232
Fukunaga K, Kawase M, Kato K (2002) Structural variation in the Waxy gene and differentiation in foxtail millet [Setaria italica (L.) P. Beauv.]: implications for multiple origins of the waxy phenotype. Mol Genet Genomics 268(2):214–222
Gao C (2021) Genome engineering for crop improvement and future agriculture. Cell 184(6):1621–1635
Gao H, Gadlage MJ, Lafitte HR, Lenderts B, Yang M, Schroder M et al (2020) Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat Biotechnol 38(5):579–581
Garcia-Huidobro T, Monteith T, Squire GR (1982) Time temperature and germination of pearl millet (Pennisetum typhoides S. & H.): I. Constant temperature. J Exp Bot 33:288–296
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 551(7681):464–471
Govindaraj M, Rai KN, Kanatti A, Upadhyaya HD, Shivade H, Rao AS (2020) Exploring the genetic variability and diversity of pearl millet core collection germplasm for grain nutritional traits improvement. Sci Rep 10(1):1–13
Gupta SK et al (2015) Seed set variability under high temperatures during flowering period in pearl millet (Pennisetum glaucum L. (R.) Br.). Field Crops Res 171:41–53
Habiyaremye C et al (2016) Proso millet (Panicum miliaceum L.) and its potential for cultivation in the Pacific northwest, US: a review. Front Plant Sci 7:1961
Holme IB, Gregersen PL, Brinch-Pedersen H (2019) Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences. Front Plant Sci 10:1468
Hunziker J et al (2020) Multiple gene substitution by target-AID base-editing technology in tomato. Sci Rep 10:20471
Ibrahim S, Saleem B, Rehman N, Zafar SA, Naeem MK, Khan MR (2021) CRISPR/Cas9 mediated disruption of inositol Pentakisphosphate 2-kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains, vol 37. J Adv Res, p 33
Jacott CN, Ridout CJ, Murray JD (2021) Unmasking mildew resistance locus O. Trends Plant Sci 26(10):1006–1013
Jain N, Arora P, Tomer R, Mishra SV, Bhatia A, Pathak H, Chakraborty D, Kumar V, Dubey D, Harit R et al (2016) Greenhouse gases emission from soils under major crops in Northwest India. Sci Total Environ 542:551–561
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Karmakar S, Das P, Panda D, Xie K, Baig MJ, Molla KA (2022) A detailed landscape of CRISPR-Cas-mediated plant disease and pest management. Plant Sci 323:111376
Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V (2019) A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565(7737):91–95
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93:1156–1160
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Kumar A, Manga VK (2011) Downy mildew of pearl millet. Biores Bull 4:182–200
Kumar S, Hash CT, Thirunavukkarasu N, Singh G, Rajaram V, Rathore A et al (2016) Mapping quantitative trait loci controlling high iron and zinc in self and open pollinated grains of pearl millet [Pennisetum glaucum (L) R. Br.]. Front Plant Sci 7:16–36
Latha AM, Rao KV, Reddy TP, Reddy VD (2006) Development of transgenic pearl millet (Pennisetum glaucum (L.) R. Br.) plants resistant to downy mildew. Plant Cell Rep 25(9):927–935
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q et al (2018) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:1–9
Li J, Wang Z, He G, Ma L, Deng XW (2020a) CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J Genet Genomics 47(5):263–272
Li Y et al (2020b) Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J 8:449–456
Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B et al (2022) Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602(7897):455–460
Liu G, Li J, Godwin ID (2019) Genome editing by CRISPR/Cas9 in sorghum through biolistic bombardment. In: Sorghum. Humana Press, New York, pp 169–183
Liu L et al (2021) Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2. Plant Biotechnol J 19:5–7
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ et al (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28(9):1998–2015
Mahendrakar MD, Parveda M, Kishor PK, Srivastava RK (2020) Discovery and validation of candidate genes for grain iron and zinc metabolism in pearl millet [Pennisetum glaucum (L.) R. Br.]. Sci Rep 10(1):1–16
Mamidi S, Healey A, Huang P, Grimwood J, Jenkins J, Barry K et al (2020) The Setaria viridis genome and diversity panel enables discovery of a novel domestication gene. bioRxiv 744557
Mariac C, Luong V, Kapran I, Mamadou A, Sagnard F, Deu M et al (2006) Diversity of wild and cultivated pearl millet accessions (Pennisetum glaucum [L.] R. Br.) in Niger assessed by microsatellite markers. Theor Appl Genet 11:49–58
Molla KA, Yang Y (2019) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37:1121–1142
Molla KA, Yang Y (2020) Predicting CRISPR/Cas9-induced mutations for precise genome editing. Trends Biotechnol 38(2):136–141
Molla KA, Qi Y, Karmakar S, Baig MJ (2020a) Base editing landscape extends to perform Transversion mutation. Trends Genet 36:899–901
Molla KA, Shih J, Yang Y (2020b) Single-nucleotide editing for zebra3 and wsl5 phenotypes in rice using CRISPR/Cas9-mediated adenine base editors. aBIOTECH 1:106–118
Molla KA, Sretenovic S, Bansal KC, Qi Y (2021) Precise plant genome editing using base editors and prime editors. Nat Plants 7(9):1166–1187
Nakayama H, Afzal M, Okuno K (1998) Intraspecific differentiation and geographical distribution of Wx alleles for low amylose content in endosperm of foxtail millet, Setaria italica (L.) Beauv. Euphytica 102(3):289–293
Nambiar VS, Dhaduk JJ, Sareen N, Shahu T, Desai R (2011) Potential functional implications of pearl millet (Pennisetum glaucum) in health and disease. J Appl Pharm Sci 1:62–67
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M et al (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353(6305):aaf8729
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T et al (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37(11):1344–1350
Oumar I et al (2008) Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br) as revealed by microsat-ellite loci. Theor Appl Genet 117:489–497
Parvathaneni RK, Spiekerman JJ, Zhou H, Wu X, Devos KM (2019) Structural characterization of ABCB1, the gene underlying the d2 dwarf phenotype in pearl millet, Cenchrus americanus (L.) Morrone. G3: Genes, Genomes. Genetics 9(8):2497–2509
Prasad PV, Staggenborg SA (2009) Growth and production of sorghum and millets. In: Soils, plant growth and crop production, vol 2. EOLSS Publishers, Oxford
Pujar M, Gangaprasad S, Govindaraj M, Gangurde SS, Kanatti A, Kudapa H (2020) Genome-wide association study uncovers genomic regions associated with grain iron, zinc and protein content in pearl millet. Sci Rep 10(1):1–15
Ramineni R, Sadumpati V, Khareedu VR, Vudem DR (2014) Transgenic pearl millet male fertility restorer line (ICMP451) and hybrid (ICMH451) expressing Brassica juncea nonexpressor of pathogenesis related genes 1 (BjNPR1) exhibit resistance to downy mildew disease. PLoS One 9(3):e90839
Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171(2):470–480
Saïdou AA, Mariac C, Luong V, Pham JL, Bezancon G et al (2009) Association studies identify natural variation at PHYC linked to flowering time and morphological variation in pearl millet. Genetics 182:899–910
Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, Barro F (2018) Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol 16(4):902–910
Sashidhar N, Harloff HJ, Potgieter L, Jung C (2020) Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol 18(11):2241–2250
Sehgal D, Skot L, Singh R, Srivastava RK, Das SP, Taunk J et al (2015) Exploring potential of pearl millet germplasm association panel for association mapping of drought tolerance traits. PLoS One 10(5):e0122165
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688
Sharma B, Chugh LK (2017) Two isoforms of lipoxygenase from mature grains of pearl millet [Pennisetum glaucum (L.) R. Br.]: purification and physico-chemico-kinetic characterization. J Food Sci Technol 54(6):1577–1584
Sharma S, Sharma R, Govindaraj M, Mahala RS, Satyavathi CT, Srivastava RK et al (2020) Harnessing wild relatives of pearl millet for germplasm enhancement: challenges and opportunities. Crop Sci 61(1):177–200
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H et al (2017) Targeted base editing in rice and tomato using a CRISPRCas9 cytidine deaminase fusion. Nat Biotechnol 35:441–443
Shivhare R, Lata C (2017) Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci Rep 6(1):1–12
Sretenovic S, Liu S, Li G, Cheng Y, Fan T, Xu Y et al (2021) Exploring C-To-G base editing in rice, tomato, and poplar. Front Genome Ed 3:756766
Tako E et al (2015) Higher iron pearl millet (Pennisetum glaucum L.) provides more absorbable iron that is limited by increased polyphenolic content. Nutr J 14(1):1–9
Vadez V, Hash T, Bidinger FR, Kholova J (2012) II 1.5 phenotyping pearl millet for adaptation to drought. Front Physiol 3:386
Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P et al (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35:969
Vinutha T, Kumar D, Bansal N, Krishnan V, Goswami S, Kumar RR et al (2022) Thermal treatments reduce rancidity and modulate structural and digestive properties of starch in pearl millet flour. Int J Biol Macromol 195:207–216
Wan DY, Guo Y, Cheng Y, Hu Y, Xiao S, Wang Y, Wen YQ (2020) CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic Res 7(1):1–14
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32(9):947–951
Wang C, Liu Q, Shen Y, Hua Y, Wang J, Lin J et al (2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol 37(3):283–286
WHO (2020) Anaemia. https://www.who.int/health-topics/anaemia#tab=tab_1
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant 6(6):1975–1983
Xing S et al (2020) Fine-tuning sugar content in strawberry. Genome Biol 21:230
Yadav AK, Arya RK, Narwal MS (2014) Screening of pearl millet F1hybrids for heat tolerance at early seedling stage. Adv Agric, Article ID. 231301. https://doi.org/10.1155/2014/231301
Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y et al (2019) Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat plants 5(5):480–485
Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y et al (2018) Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36:950–953
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Panda, D., Baig, M.J., Molla, K.A. (2024). Genome Editing and Opportunities for Trait Improvement in Pearl Millet. In: Tonapi, V.A., Thirunavukkarasu, N., Gupta, S., Gangashetty, P.I., Yadav, O. (eds) Pearl Millet in the 21st Century. Springer, Singapore. https://doi.org/10.1007/978-981-99-5890-0_7
Download citation
DOI: https://doi.org/10.1007/978-981-99-5890-0_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-5889-4
Online ISBN: 978-981-99-5890-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)