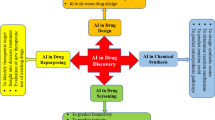
Abstract
Pharmaceutical companies and chemical experts invest much in drug design and development research. Low effectiveness, off-target delivery, higher costs, and time intake present barriers and constraints to drug design and development. Furthermore, complicated and extensive data sets generated from genomes, proteomics, microarray data, and clinical trials obstruct drug development as recorded data are tough to analyze and model. Artificial intelligence (AI) has been widely applied in drug research and has contributed significantly to its design and development. Deep learning, a subdomain of AI, has revolutionized the field of drug discovery, especially for peptide synthesis, toxicity prediction, drug repositioning, etc. In this chapter, numerous application areas have been identified where existing AI technologies have the potential to speed up drug design research work. Recent progress in the AI field has opened new horizons for drug discovery research. This chapter discusses the current approaches, technological obstacles, and limitations with the goal of probable future avenues for AI-aided drug development and discovery.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Eppe M., Nguyen, P.D., Wermter, S.: From semantics to execution: Integrating action planning with reinforcement learning for robotic causal problem-solving. Front. Robot. AI 6(123), (2019)
Hamet, P., Tremblay, J.: Artificial intelligence in medicine. Metabolism 69, S36–S40 (2017)
Duch, W., Swaminathan, K., Meller, J.: Artificial intelligence approaches for rational drug design and discovery. Curr. Pharm. Des. 13(14), 1497–1508 (2007)
Fukushima, K.: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 36(4), 193–202 (1980)
Kim, S., Chen, J., Cheng, T., Gindulyte., A., He, J., He, S., Bolton, E.E.: PubChem in 2021: new data content and improved web interfaces. Nucl. Acids Res. 49, 1388–1395 (2021)
Baldi, A.: Computational approaches for drug design and discovery: an overview. Syst. Rev. Pharm. 1(1), 99 (2010)
Kalaiarasi, C., Manjula, S., Kumaradhas, P.: Combined quantum mechanics/molecular mechanics (QM/MM) methods to understand the charge density distribution of estrogens in the active site of estrogen receptors. RSC Adv. 9(69), 40758–40771 (2019)
Carpenter, K.A., Huang, X.: Machine learning-based virtual screening and its applications to Alzheimer’s drug discovery: a review. Curr. Pharm. Des. 24(28), 3347–3358 (2018)
Provenzano, C., Cappella, M., Valaperta, R., Cardani, R., Meola, G., Martelli, F., Falcone, G.: CRISPR/Cas9-mediated deletion of CTG expansions recovers normal phenotype in myogenic cells derived from myotonic dystrophy 1 patients. Molecular Therapy-Nucleic Acids 9, 337–348 (2017)
Mustapha, I.B., Saeed, F.: Bioactive molecule prediction using extreme gradient boosting. Molecules 21(8), 983 (2016)
Drouin, A., Letarte, G., Raymond, F., Marchand, M., Corbeil, J., Laviolette, F.: Interpretable genotype-to-phenotype classifiers with performance guarantees. Sci. Rep. 9(1), 4071 (2019). https://doi.org/10.1038/s41598-019-40561-2
Ramon, E., Belanche-Muñoz, L., Pérez-Enciso, M.: HIV drug resistance prediction with weighted categorical kernel functions. BMC Bioinform. 20(1), 1–13 (2019)
Chen, M.L., et al.: Beyond multidrug resistance: leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction. EBioMedicine 43, 356–369 (2019). https://doi.org/10.1016/j.ebiom.2019.04.016
Chen, T., Guestrin, C.: Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794 (2016).
Rishishwar, L., Petit, R.A., Kraft, C.S., Jordan, I.K.: Genome sequence-based discriminator for vancomycin-intermediate Staphylococcus aureus. J. Bacteriol. 196(5), 940–948 (2014). https://doi.org/10.1128/JB.01410-13
Coelho, J.R., et al.: The use of machine learning methodologies to analyse antibiotic and biocide susceptibility in Staphylococcus aureus. PLoS ONE 8(2), e55582 (2013). https://doi.org/10.1371/journal.pone.0055582
Goodman, K.E., Lessler, J., Harris, A.D., Milstone, A.M., Tamma, P.D.: A methodological comparison of risk scores versus decision trees for predicting drug-resistant infections: a case study using extended-spectrum beta-lactamase (ESBL) bacteremia. Infect. Control Hosp. Epidemiol. 40(4), 400–407 (2019). https://doi.org/10.1017/ice.2019.17
Raposo, L.M., Arruda, M.B., de Brindeiro, R.M., Nobre, F.F.: Lopinavir resistance classification with imbalanced data using probabilistic neural networks. J. Med. Syst. 40(3), 69 (2016). https://doi.org/10.1007/s10916-015-0428-7
Bhattacharyya, R.P., et al.: Simultaneous detection of genotype and phenotype enables rapid and accurate antibiotic susceptibility determination. Nat. Med. 25(12), 1858–1864 (2019). https://doi.org/10.1038/s41591-019-0650-9
Sauer, C.M., et al.: Feature selection and prediction of treatment failure in tuberculosis. PLoS ONE 13(11), e0207491 (2018). https://doi.org/10.1371/journal.pone.0207491
Wicht, K.J., Combrinck, J.M., Smith, P.J., Egan, T.J.: Bayesian models trained with HTS data for predicting β-haematin inhibition and in vitro antimalarial activity. Bioorg. Med. Chem. 23(16), 5210–5217 (2015)
Rogers, D., Brown, R.D., Hahn, M.: Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J. Biomol. Screen. 10(7), 682–686 (2005)
Speck-Planche, A.V., Kleandrova, V., Luan, F., Cordeiro, N.D.: Chemoinformatics in multi-target drug discovery for anti-cancer therapy: in silico design of potent and versatile anti-brain tumor agents. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 12(6), 678 (2012).
Xia, X., Maliski, E.G., Gallant, P., Rogers, D.: Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 47(18), 4463–4470 (2014)
Ouyang, X., Handoko, S.D., Kwoh, C.K.: Cscore: a simple yet effective scoring function for protein–ligand binding affinity prediction using modified cmac learning architecture. J. Bioinform. Comput. Biol. 9(1), 1–14 (2011)
Srivastava, R.K., Greff, K., Schmidhuber, J.: Training very deep networks. Adv. Neural Inf. Process. Syst., 28 (2015).
Liu, B., Ramsundar, B., Kawthekar, P., Shi, J., Gomes, J., LuuNguyen, Q., Pande, V.: Retrosynthetic reaction prediction using neural sequence-to-sequence models. ACS Cent. Sci. 3(10), 1103–1113 (2017)
Schneider, G., Clark, D.E.: Automated de novo drug design: are we nearly there yet. Angew. Chem. Int. Ed. 58(32), 10792–10803 (2019)
Asanuma, D., Sakabe, M., Kamiya, M., Yamamoto, K., Hiratake, J., Ogawa, M., Urano, Y.: Sensitive β-galactosidase-targeting fluorescence probe for visualizing small peritoneal metastatic tumours in vivo. Nat. Commun. 6(1), 1–7 (2015)
Jain, A., Zamir, A.R., Savarese, S., Saxena, A.: Structural-rnn: Deep learning on Spatio-temporal graphs. In Proceedings of the ieee conference on computer vision and pattern recognition, 5308–5317(2016)
Sanchez-Lengeling, B., Aspuru-Guzik, A.: Inverse molecular design using machine learning: Generative models for matter engineering. Science 361(6400), 360–365 (2018)
Sellers, B.D., James, N.C., Gobbi, A.: A comparison of quantum and molecular mechanical methods to estimate strain energy in druglike fragments. J. Chem. Inf. Model. 57(6), 1265–1275 (2017)
Popova, M., Isayev, O., Tropsha, A.: Deep reinforcement learning for de novo drug design. Sci. Adv. 4(7), 7885 (2018)
Segler, M.H., Preuss, M., Waller, M.P.: Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555(7698), 604–610 (2018)
Li, L., Snyder, J.C., Pelaschier, I.M., Huang, J., Niranjan, U.N., Duncan. P., Burke, K. Understanding machine‐learned density functionals. Int. J. Quantum Chem. 116(11), 819–833 (2016).
Pilania, A., Mannodi-Kanakkithodi, B.P., Uberuaga, R., Ramprasad, J.E., Gubernatis, Lookman, T.: Machine learning bandgaps of double perovskites. Sci. Rep. 6, 19375 (2016).
Pilania, G., Mannodi-Kanakkithodi, A., Uberuaga, B., et al.: Machine learning bandgaps of double perovskites. Sci. Rep. 6, 19375 (2016)
Margolis, R., Derr, L., Dunn, M., Huerta, M., Larkin, J., Sheehan. J,, Green, E.D.: The National Institutes of Health's Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J. Am. Med. Inform. Assoc. 21(6), 957–958 (2014).
Parmar, C., Barry, J.D., Hosny, A., Quackenbush, J., Aerts, H.J.: Data analysis strategies in medical imagingData science designs in medical imaging. Clin. Cancer Res. 24(15), 3492–3499 (2018)
Cohen, J.D., Li, L., Wang, Y., Thoburn, C., Afsari, B., Danilova, L., Papadopoulos, N.: Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359(6378), 926–930 (2018)
Wang, H.-Y., et al.: Rapid Detection of Heterogeneous Vancomycin-Intermediate Staphylococcus aureus Based on Matrix-Assisted Laser Desorption Ionization Time-of-Flight: Using a Machine Learning Approach and Unbiased Validation. Front. Microbiol. 9, 2393 (2018). https://doi.org/10.3389/fmicb.2018.02393
Huang, T.-S., Lee, S.S.-J., Lee, C.-C., Chang, F.-C.: Detection of carbapenem-resistant Klebsiella pneumoniae on the basis of matrix-assisted laser desorption ionization time-offlight mass spectrometry by using supervised machine learning approach. PLoS ONE 15(2), e0228459 (2020). https://doi.org/10.1371/journal.pone.0228459
Zhang, C., et al.: Systematic analysis of supervised machine learning as an effective approach to predicate β-lactam resistance phenotype in Streptococcus pneumoniae. Brief. Bioinform. 21(4), 1347–1355 (2020). https://doi.org/10.1093/bib/bbz056
Moradigaravand, D., Palm, M., Farewell, A., Mustonen, V., Warringer, J., Parts, L.: Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput. Biol. 14(12), e1006258 (2018). https://doi.org/10.1371/journal.pcbi.1006258
Feretzakis, G., et al.: Using machine learning techniques to aid empirical antibiotic therapy decisions in the intensive care unit of a general hospital in Greece. Antibiot. Basel Switz. 9(2) (2020). https://doi.org/10.3390/antibiotics9020050.
Haga, H., et al.: A machine learning-based treatment prediction model using whole genome variants of hepatitis C virus. PLoS ONE 15(11), e0242028 (2020). https://doi.org/10.1371/journal.pone.0242028
Oonsivilai, M., et al.: Using machine learning to guide targeted and locally-tailored empiric antibiotic prescribing in a children’s hospital in Cambodia. Wellcome Open Res. 3, 131 (2018). https://doi.org/10.12688/wellcomeopenres.14847.1
Macesic, N.N., Bear Don’t Walk, O.J., Pe’er, I., Tatonetti, N.P., Peleg, A.Y., Uhlemann, A.-C.: Predicting phenotypic Polymyxin Resistance in Klebsiella pneumoniae through Machine Learning Analysis of Genomic Data. mSystems 5(3) (2020). https://doi.org/10.1128/mSystems.00656-19
Kouchaki, S., et al.: Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinforma. Oxf. Engl. 35(13), 2276–2282 (2019). https://doi.org/10.1093/bioinformatics/bty949
Mason, D.J., et al.: Prediction of Antibiotic Interactions Using Descriptors Derived from Molecular Structure. J. Med. Chem. 60(9), 3902–3912 (2017). https://doi.org/10.1021/acs.jmedchem.7b00204
Gupta, R., Srivastava, D., Sahu, M., et al.: Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers 25, 1315–1360 (2021). https://doi.org/10.1007/s11030-021-10217-3
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Nayan, K., Paswan, K.K., Sharma, V.B., Kumar, Y., Tewari, S. (2023). Recent Advancements in AI-Assisted Drug Design and Discovery Systems. In: Mishra, A., Lin, J.CW. (eds) Industry 4.0 and Healthcare . Advanced Technologies and Societal Change. Springer, Singapore. https://doi.org/10.1007/978-981-99-1949-9_2
Download citation
DOI: https://doi.org/10.1007/978-981-99-1949-9_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-1948-2
Online ISBN: 978-981-99-1949-9
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)