Abstract
Machine learning (ML) is the most promising subset of artificial intelligence. Quantum computing is prevalent for fast problem-solving approaches. The complex problems are classified and solved using huge multi-dimensional space. The various algorithms can interfere in multi-dimensional space and resolve the problems. Quantum Machine Learning provides the platform for various mining processes with to the point developments in quantum computing. Quantum computing & Machine learning both are very complex. Quantum Machine learning focuses on quick problem-solving synthesis with a quantum framework using different algorithms. Machine Learning functions by supervised, unsupervised, and semi-supervised learning mechanisms. ML uses label and unlabeled data to implement different classification, clustering, and decision trees for complex problems. Quantum computing comprises quantum counterparts for various computational complexity. Quantum Machine Learning provides a profound sympathetic approach for various subjects to derive new dimensioned results. There are several serious life-threatening diseases such as cancer, hepatotoxicity, cardiotoxicity, nephrotoxicity, etc. require prompt and precise detection at the early stages of progression. The need of the hour is to develop rapid, accurate, and more efficient strategies for various disease predictions which are also cost-effective and non-invasive in nature. Breast cancer is also such a disease that early screening is challenging owning to hereditary predisposition. Quantum computation techniques emerged with Machine learning as the promising approach in the past decade concerning the prediction of breast cancer. The quantum computes can be utilized for assisting cancer detection by employing quantum neural networks, quantum simulators, Super Vector Machine (SVM); Artificial Neural Networks (ANN), Dimensionality Reduction Algorithms etc. are used on the pre-processed dataset for the derived prediction of breast cancer. This book chapter will focus on current trends of Quantum Machine leaning for the prediction of breast cancers by solving complex computational problems using above stated algorithms. This chapter discusses the Molecular Classification of Breast Cancer as Luminal-A, Luminal-B, Normal-like, HER2 enriched, and Basal-like with Breast Cancer Diagnostic Techniques. It covers the study of Brest cancer prediction using Quantum Neural Network, Dimensionality Reduction Algorithms, and Support vector machines (SVM). It includes comparative discussions about different algorithms for breast cancer prediction.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
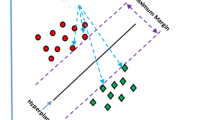
Similar content being viewed by others
References
D. Quiroga, P. Date, R. Pooser, Discriminating quantum states with quantummachine learning, in 2021 IEEE International Conference on Quantum Computing and Engineering (QCE), 17–22 October 2021, pp. 481–482
M. Nivelkar, S.G. Bhirud, Optimized machine learning: training and classification performance using quantum computing, in 2021 IEEE 6th International Conference on Computing, Communication and Automation (ICCCA), 17–19 December 2021, pp. 8–13
P. Sangeetha, P. Kumari, Quantumalgorithms for machine learning and optimization, in 2020 2nd PhD Colloquium on Ethically Driven Innovation and Technology for Society (PhD EDITS), 8 November 2020, pp. 1–2
P. Rebentrost, M. Mohseni, S. Lloyd, Quantum support vector machine for big data classification. Phys. Rev. Lett. 113(13), 130503 (2014). https://doi.org/10.1103/PhysRevLett.113.130503
G. Bonaccorso, Machine Learning Algorithms (Packt Publishing Ltd., 2017)
F. Schwenker, E. Trentin, Pattern classification and clustering: a review of partially supervised learning approaches. Pattern Recogn. Lett. 37, 4–14 (2014). https://doi.org/10.1016/j.patrec.2013.10.017
M. Riedmiller, Advanced supervised learning in multi-layer perceptrons—from backpropagation to adaptive learning algorithms. Comput. Stand. Interf. 16(3), 265–278 (1994)
S.S. Sawant, M. Prabukumar, A review on graph-based semi-supervised learning methods for hyperspectral image classification. Egypt. J. Remote Sens. Space Sci. 23(2), 243–248 (2020). https://doi.org/10.1016/j.ejrs.2018.11.001
N. Li, M. Shepperd, Y. Guo, A systematic review of unsupervised learning techniques for software defect prediction. Inf. Softw. Technol. 122, 106287 (2020). https://doi.org/10.1016/j.infsof.2020.106287
T.O. Ayodele, Types of machine learning algorithms. New Adv. Mach. Learn. 3, 19–48 (2010)
R. Chatterjee, T. Yu, Generalized coherent states, reproducing kernels, and quantum support vector machines (2016). arXiv preprint arXiv:1612.03713
Z. Zhao, J.K. Fitzsimons, J.F. Fitzsimons, Quantum-assisted Gaussian process regression. Phys. Rev. A 99(5), 052331 (2019)
J.D. Whitfield, M. Faccin, J.D. Biamonte, Ground-state spin logic. EPL (Europhys. Lett.) 99(5), 57004 (2012)
Y. Bengio, Y. LeCun, Scaling learning algorithms towards AI. Large-Scale Kernel Mach. 34(5), 1–41 (2007)
M. Denil,N. De Freitas, Toward the implementation of a quantum RBM (2011)
V. Dumoulin, I. Goodfellow, A. Courville, Y. Bengio, On the challenges of physical implementations of RBMs’ (2014)
J. Biamonte, P. Wittek, N. Pancotti, P. Rebentrost, N. Wiebe, S. Lloyd, Quantum machine learning. Nature 549(7671), 195–202 (2017). https://doi.org/10.1038/nature23474
J.D. Biamonte, P.J. Love, Realizable Hamiltonians for universal adiabatic quantum computers. Phys. Rev. A 78(1), 012352 (2008)
S. Lloyd, B.M. Terhal, Adiabatic and Hamiltonian computing on a 2D lattice with simple two-qubit interactions. New J. Phys. 18(2), 023042 (2016)
S.-Y. Xia, Z.-Y. Xiong, Y.-G. Luo, L.-M. Dong, A method to improve support vector machine based on distance to hyperplane, 126(20), 2405–2410 (2015)
E. Farhi, J. Goldstone, S. Gutmann, A quantum approximate optimization algorithm (2014). arXiv preprint arXiv:1411.4028
S. Lloyd, M. Mohseni, P. Rebentrost, Quantum algorithms for supervised andunsupervised machine learning (2013). arXiv preprint arXiv:1307.0411
J. Schmidhuber, Deep learning in neural networks: an overview. Neural Netw. 61, 85–117 (2015). https://doi.org/10.1016/j.neunet.2014.09.003
S. Lloyd, C. Weedbrook, Quantum generative adversarial learning. Phys. Rev. Lett. 121(4), 040502 (2018). https://doi.org/10.1103/PhysRevLett.121.040502
I. Cong, S. Choi, M.D. Lukin, Quantum convolutional neural networks. Nat. Phys. 15(12), 1273–1278 (2019). https://doi.org/10.1038/s41567-019-0648-8
C. Lavor, L.R.U. Manssur, R. Portugal, Grover’s algorithm: Quantum database search (2003). arXiv preprint quant-ph/0301079
M. Kaloev, G. Krastev, Experiments focused on exploration in deep reinforcement learning, in 2021 5th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), 21–23 October 2021, pp. 351–355
W.D. Smart, L.P. Kaelbling, Effective reinforcement learning for mobile robots, in Proceedings 2002 IEEE International Conference on Robotics and Automation (Cat. No. 02CH37292) (IEEE, 2002), pp. 3404–3410
E. Aïmeur, G. Brassard, S. Gambs, Quantum speed-up for unsupervised learning. Mach. Learn. 90(2), 261–287 (2013). https://doi.org/10.1007/s10994-012-5316-5
A. Bisio, G. Chiribella, G.M. d Ariano, S. Facchini, P. Perinotti, Optimal quantum learning of a unitary transformation, 81, 032324 (2010)
S. Lloyd, M. Mohseni, P. Rebentrost, Quantum principal componentanalysis, 10, 631–633 (2014)
D. Ventura, T. Martinez, Quantum associative memory. Inf. Sci. 124(1), 273–296 (2000). https://doi.org/10.1016/S0020-0255(99)00101-2
N. Wiebe, A. Kapoor, K. Svore, Quantum algorithms for nearest-neighbor methods for supervised and unsupervised learning (2014). arXiv preprint arXiv:1401.2142
A. Narayanan, T. Menneer, Quantum artificial neural network architectures and components. Inform. Sci. 128, 231–255 (2000a). https://doi.org/10.1016/S0020-0255(00)00055-4
D. Anguita, S. Ridella, F. Rivieccio, R. Zunino, Quantum optimization for training support vector machines. Neural Netw. 16(5–6), 763–770 (2003). https://doi.org/10.1016/s0893-6080(03)00087-x
K.-A. Brickman, P. Haljan, P. Lee, M. Acton, L. Deslauriers, C. Monroe, Implementation of Grover’s quantum search algorithm in a scalable system, 72(5), 050306 (2005)
P. Kwiat, J. Mitchell, P. Schwindt, A.G. White, Grover’s search algorithm: an optical approach, 47(2–3), 257–266 (2000)
C. Allauzen, M. Crochemore, M. Raffinot, Factor oracle: a new structure for pattern matching, in International Conference on Current Trends in Theory and Practice of Computer Science (Springer, 1999), pp. 295–310
D. Dong, C. Chen, H. Li, T. Tarn, Quantum reinforcement learning. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 38(5), 1207–1220 (2008). https://doi.org/10.1109/TSMCB.2008.925743
K. Kashyap, S.D. Lalit, L. Gautam, From classical to quantum: a review ofrecent progress in reinforcement learning, in 2021 2nd International Conference for Emerging Technology (INCET). 21–23 May 2021, pp. 1–5
C. Chen, D. Dong, Complexity analysis of Quantum reinforcement learning, in Proceedings of the 29th Chinese Control Conference, 29–31 July 2010, pp. 5897–5901
A.J. Smith, Applications of the self-organising map to reinforcement learning. Neural Netw. 15(8–9), 1107–1124 (2002)
C. Chen, H.-X. Li, D. Dong, Hybrid control for robotnavigation-a hierarchical q-learning algorithm, 15(2), 37–47 (2008)
M. Kaya, R. Alhajj, A novel approach to multiagent reinforcement learning: utilizing OLAP mining in the learning process. IEEE Trans. Syst. Man Cybern. Part C (Appl. Rev.) 35(4), 582–590. https://doi.org/10.1109/TSMCC.2004.843188
S. Whiteson, Evolutionary function approximation for reinforcement learning, 7 (2006)
T. Hogg, D. Portnov, Quantum optimization. Inf. Sci. 128(3–4), 181–197 (2000)
A.A. Ezhov, D. Ventura, Quantum neural networks, in Future Directions for Intelligent Systems and Information Sciences (Springer, 2000), pp. 213–235
C.M. Catherine, Adiabatic Quantum Computation and Quantum Annealing: Theory and Practice (Morgan & Claypool, 2014)
Y. Matsuda, H. Nishimori, H.G. Katzgraber, Ground-state statistics from annealing algorithms: quantum versus classical approaches. New J. Phys. 11(7), 073021 (2009)
G.E. Santoro, R. Martonák, E. Tosatti, R. Car, Theory of quantum annealing of an Ising spin glass. Science 295(5564), 2427–2430 (2002)
W. Vinci,K. Markström, S. Boixo, A. Roy, F.M. Spedalieri, P.A. Warburton, et al., Hearing the shape of the Ising model with a programmable superconducting-flux annealer, 4(1), 1–7 (2014)
N. Chancellor, S. Szoke, W. Vinci, G. Aeppli, P.A. Warburton, Maximum- entropy inference with a programmable annealer, 6(1), 1–14 (2016)
Y. Otsubo, J.-I. Inoue, K. Nagata, M. Okada, Effect of quantumfluctuation in error-correcting codes, 86(5), 051138 (2012)
Y. Otsubo, J.-I. Inoue, K. Nagata, M. Okada, Code-division multiple-access multiuser demodulator by using quantum fluctuations, 90(1), 012126 (2014)
V. Choi, Adiabatic quantum algorithms for the NP-complete maximum—weightindependent set, exact cover and 3SAT problems (2010)
S.H. Adachi, M.P. Henderson, Application of quantum annealing to the training of deep neural networks (2015). arXiv preprint arXiv:1510.06356
M.H. Amin, E. Andriyash, J. Rolfe, B. Kulchytskyy, R. Melko, Quantum Boltzmann machine. Phys. Rev. X 8(2), 021050 (2018)
K. Hukushima, K. Nemoto, Exchange Monte Carlo method and application to spin glass simulations, 65(6), 1604–1608 (1996)
S. Kirkpatrick, C.D. Gelatt Jr, M.P. Vecchi, Optimization by simulatedannealing, 220(4598), 671–680 (1983)
D. de Falco, D. Tamascelli, An introduction to quantum annealing, 45(1), 99–116 (2011)
Morita, S., & Nishimori, H (2008). Mathematical foundation of quantum annealing. 49(12), 125210
M. Panella, G. Martinelli, Neural networks with quantum architecture and quantum learning. Int. J. Circ. Theory Appl. 39(1), 61–77 (2011)
R. Zhou, H. Zheng, N. Jiang, Q. Ding, Self-organizing Quantum Neural Network (IEEE, 2006), pp. 1067–1072
A.J. da Silva, T.B. Ludermir, W.R. de Oliveira, Quantum perceptron over a field and neural network architecture selection in a quantum computer. Neural Netw. 76, 55–64 (2016)
S.C. Kak, Quantum neural computing. Adv. Imag. Electron. Phys. 94, 259–313 (1995)
A. Narayanan,T. Menneer, Quantum artificial neural network architectures and components, 128(3–4), 231–255 (2000b)
S. Ding, Z. Zhu, X. Zhang, An overview on semi-supervised support vector machine. Neural Comput. Appl. 28(5), 969–978 (2017)
S. Ding, X. Hua, J. Yu, An overview on nonparallel hyperplane support vector machine algorithms. Neural Comput. Appl. 25(5), 975–982 (2014). https://doi.org/10.1007/s00521-013-1524-6
Y.-H. Shao,W.-J. Chen, N.-Y. Deng, Nonparallel hyperplane support vector machine for binary classification problems. Inform. Sci. 263, 22–35 (2014). https://doi.org/10.1016/j.ins.2013.11.003
H. Xu, C. Guedes Soares, Manoeuvring modelling of a containership in shallowwater based on optimal truncated nonlinear kernel-based least square support vector machine and quantum-inspired evolutionary algorithm. Ocean Eng. 195, 106676 (2020). https://doi.org/10.1016/j.oceaneng.2019.106676
Y. Zhang, Q. Ni, Recent advances in quantum machine learning, 2(1), e34 (2020). https://doi.org/10.1002/que2.34
D.M. Hausman, What is cancer? Perspect Biol Med 62(4), 778–784 (2019). https://doi.org/10.1353/pbm.2019.0046
P.S. Roy, B.J. Saikia, Cancer and cure: a critical analysis. Indian J. Cancer 53(3), 441–442 (2016). https://doi.org/10.4103/0019-509x.200658
R.L. Siegel, K.D. Miller, H.E. Fuchs, A. Jemal, Cancer statistics, 72(1), 7– 33 (2022). https://doi.org/10.3322/caac.21708
M.A. Zaimy, N. Saffarzadeh, A. Mohammadi, H. Pourghadamyari, P. Izadi, A. Sarli et al., New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene. Ther. 24(6), 233–243 (2017). https://doi.org/10.1038/cgt.2017.16
P. Mathur, K. Sathishkumar, M. Chaturvedi, P. Das, K.L. Sudarshan, S. Santhappan, et al., Cancer Statistics, 2020: Report From National Cancer Registry Programme, India, (6), 1063-1075 (2020). https://doi.org/10.1200/go.20.00122
R. Fisher, L. Pusztai, C. Swanton, Cancer heterogeneity: implications for targeted therapeutics. Br. J. Cancer 108(3), 479–485 (2013). https://doi.org/10.1038/bjc.2012.581
C.E. Meacham, S.J. Morrison, Tumour heterogeneity and cancer cell plasticity. Nature 501(7467), 328–337 (2013). https://doi.org/10.1038/nature12624
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 66(1), 7–30. (2016). https://doi.org/10.3322/caac.21332
K. Aizawa, C. Liu, S. Tang, S. Veeramachaneni, K.-Q. Hu, D.E. Smith, et al., Tobacco carcinogen induces both lung cancer and non-alcoholic steatohepatitis and hepatocellular carcinomas in ferrets which can be attenuated by lycopene supplementation, 139(5), 1171–1181 (2016). https://doi.org/10.1002/ijc.30161
S.O. Antwi, E.C. Eckert, C.V. Sabaque, E.R. Leof, K.M. Hawthorne, W.R. Bamlet et al., Exposure to environmental chemicals and heavy metals, and risk of pancreatic cancer. Cancer Causes & Control 26(11), 1583–1591 (2015). https://doi.org/10.1007/s10552-015-0652-y
M.G.K. Cumberbatch, A. Cox, D. Teare, J.W.F. Catto, Contemporary occupational carcinogen exposure and bladder cancer: a systematic review and meta-analysis. JAMA Oncol. 1(9), 1282–1290 (2015). https://doi.org/10.1001/jamaoncol.2015.3209%JJAMAOncology
S.L. Poon, J.R. McPherson, P. Tan, B.T. Teh, S.G. Rozen, Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Gen. Med. 6(3), 24 (2014). https://doi.org/10.1186/gm541
D.M. Parkin, The global health burden of infection-associated cancers in the year, 118(12), 3030–3044 (2006). https://doi.org/10.1002/ijc.21731
Z. Anastasiadi, G.D. Lianos, E. Ignatiadou, H.V. Harissis, M. Mitsis, Breast cancer in young women: an overview. Updat. Surg. 69(3), 313–317 (2017). https://doi.org/10.1007/s13304-017-0424-1
A. Metelková, A. Skálová, J. Fínek, Breast cancer in young women-correlation of clinical histomorphological, and molecular-genetic features of breast carcinoma in women younger than 35 years of age. Klin Onkol 30(3), 202–209 (2017). https://doi.org/10.14735/amko2017202
H.A. Azim, A.H. Partridge, Biology of breast cancer in young women. Breast Cancer Res. 16(4), 427 (2014). https://doi.org/10.1186/s13058-014-0427-5
F.K. Al-Thoubaity, Molecular classification of breast cancer: A retrospective cohort study. Ann. Med. Surg. (Lond.) 49, 44–48. https://doi.org/10.1016/j.amsu.2019.11.021
N. Eliyatkın, E. Yalçın, B. Zengel, S. Aktaş, E. Vardar, Molecular classification of breast carcinoma: from traditional, old-fashioned way to a new age, and a new way. J. Breast Health 11(2), 59–66 (2015). https://doi.org/10.5152/tjbh.2015.1669
E.A. Rakha, I.O. Ellis, Modern classification of breast cancer: should we stick with morphology or convert to molecular profile characteristics. Adv. Anat. Pathol. 18(4) 255–267 (2011). https://doi.org/10.1097/PAP.0b013e318220f5d1
G. Viale, The current state of breast cancer classification. Ann. Oncol. 23, x207–x210 (2012). https://doi.org/10.1093/annonc/mds326
A. Blagodatski, V. Cherepanov, A. Koval, V.I. Kharlamenko, Y.S. Khotimchenko, V.L. Katanaev, High-throughput targeted screening in triple-negative breast cancer cells identifies Wnt-inhibiting activities in Pacific brittle stars. Sci. Rep. 7(1), 11964 (2017). https://doi.org/10.1038/s41598-017-12232-7
P.A. Fitzpatrick, N. Akrap, E.M.V. Söderberg, H. Harrison, G.J. Thomson, G. Landberg, Robotic mammosphere assay for high-throughput screening in triple-negative breast cancer. SLAS Discov. 22(7), 827–836 (2017). https://doi.org/10.1177/2472555217692321
L. Huang, X. Yi, X. Yu, Y. Wang, C. Zhang, L. Qin, et al., High-throughput strategies for the discovery of anticancer drugs by targeting transcriptional reprogramming (review), 11 (2021). https://doi.org/10.3389/fonc.2021.762023
Z. Sun, B. Zhang, High-throughput screening (HTS) of natural products with triple- negative breast cancer (TNBC) organoids, 37(15_suppl), e12558 (2019). https://doi.org/10.1200/JCO.2019.37.15_suppl.e12558
F.K. Al-thoubaity, Molecular classification of breast cancer: a retrospective cohort study. Ann. Med. Surg. 49, 44–48 (2020a). https://doi.org/10.1016/j.amsu.2019.11.021
H.O. Habashy, D.G. Powe, T.M. Abdel-Fatah, J.M. Gee, R.I. Nicholson, A.R. Green et al., A review of the biological and clinical characteristics of luminal-like oestrogen receptor-positive breast cancer. Histopathology 60(6), 854–863 (2012). https://doi.org/10.1111/j.1365-2559.2011.03912.x
C. Sotiriou, S.-Y. Neo, L.M. McShane, E.L. Korn, P.M. Long, A. Jazaeri et al., Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. U.S.A. 100(18), 10393–10398 (2003). https://doi.org/10.1073/pnas.1732912100
L.A. Carey, Through a glass darkly: advances in understanding breast cancer biology, 2000–2010. Clin Breast Cancer 10(3), 188–195 (2010). https://doi.org/10.3816/CBC.2010.n.026
J.J. Gao, S.M. Swain, Luminal a breast cancer and molecular assays: a review. Oncologist 23(5), 556–565 (2018). https://doi.org/10.1634/theoncologist.2017-0535
V. Guarneri, P. Conte, Metastatic breast cancer: therapeutic options according to molecular subtypes and prior adjuvant therapy. Oncologist 14(7), 645–656 (2009). https://doi.org/10.1634/theoncologist.2009-0078
H. Kennecke, R. Yerushalmi, R. Woods, M.C. Cheang, D. Voduc, C.H. Speers et al., Metastatic behavior of breast cancer subtypes. J Clin Oncol 28(20), 3271–3277 (2010). https://doi.org/10.1200/jco.2009.25.9820
S. Dwivedi, P. Purohit, R. Misra, M. Lingeswaran, J.R. Vishnoi, P. Pareek, et al.,Chapter 5—Application of single-cell omics in breast cancer, in Single-Cell Omics, ed. by D. Barh, V. Azevedo (Academic Press, 2019), pp. 69–103. https://doi.org/10.1016/B978-0-12-817532-3.00005-0
Z.-H. Li, P.-H. Hu, J.-H. Tu, N.-S. Yu, Luminal B breast cancer: patterns of recurrence and clinical outcome. Oncotarget 7(40), 65024–65033 (2016). https://doi.org/10.18632/oncotarget.11344
B. Tran, P.L. Bedard, Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 13(6), 221 (2011). https://doi.org/10.1186/bcr2904
M.J. Ellis, Y. Tao, J. Luo, R. A’Hern, D.B. Evans, A.S. Bhatnagar et al., Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J. Natl. Cancer Inst. 100(19), 1380–1388 (2008). https://doi.org/10.1093/jnci/djn309
R. Bhargava, S. Beriwal, D.J. Dabbs, U. Ozbek, A. Soran, R.R. Johnson et al., Immunohistochemical surrogate markers of breast cancer molecular classes predicts response to neoadjuvant chemotherapy: a single institutional experience with 359 cases. Cancer 116(6), 1431–1439 (2010). https://doi.org/10.1002/cncr.24876
S. Paik, S. Shak, G. Tang, C. Kim, J. Baker, M. Cronin et al., A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351(27), 2817–2826 (2004). https://doi.org/10.1056/NEJMoa041588
J.S. Parker, A. Prat, M. Cheang, M. Lenburg, S. Paik, C.J.C.R. Perou, Breast cancer molecular subtypes predict response to anthracycline/taxane-based chemotherapy, 69(24 Suppl 3) (2009)
B. Weigelt, A. Mackay, R. A’Hern, R. Natrajan, D.S. Tan, M. Dowsett et al., Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 11(4), 339–349 (2010). https://doi.org/10.1016/s1470-2045(10)70008-5
L. Yin, J.-J. Duan, X.-W. Bian, S.-C. Yu, Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22(1), 61 (2020). https://doi.org/10.1186/s13058-020-01296-5
J.J. de Ronde, J. Hannemann, H. Halfwerk, L. Mulder, M.E. Straver, M.J. Vrancken Peeters, et al., Concordance of clinical and molecular breast cancer subtyping in the context of preoperative chemotherapy response. Breast Cancer Res. Treat 119(1), 119–126 (2010). https://doi.org/10.1007/s10549-009-0499-6
U. Krishnamurti, J.F. Silverman, HER2 in breast cancer: a review and update. Adv. Anat. Pathol. 21(2) (2014). https://journals.lww.com/anatomicpathology/Fulltext/2014/03000/HER2_in_Breast_Cancer__A_Review_and_Update.4.aspx
J.S. Ross, J.A. Fletcher, G.P. Linette, J. Stec, E. Clark, M. Ayers et al., The Her-2/neu gene and protein in breast cancer 2003: biomarker and target of therapy. Oncologist 8(4), 307–325 (2003). https://doi.org/10.1634/theoncologist.8-4-307
J. Wang, B. Xu, Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Signal Transduct. Target. Ther. 4(1), 34 (2019). https://doi.org/10.1038/s41392-019-0069-2
B. Kreike, M. van Kouwenhove, H. Horlings, B. Weigelt, H. Peterse, H. Bartelink et al., Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res 9(5), R65 (2007). https://doi.org/10.1186/bcr1771
E.A. Rakha, S.E. Elsheikh, M.A. Aleskandarany, H.O. Habashi, A.R. Green, D.G. Powe et al., Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin. Cancer Res. 15(7), 2302–2310 (2009). https://doi.org/10.1158/1078-0432.Ccr-08-2132
E.R. Myers, P. Moorman, J.M. Gierisch, L.J. Havrilesky, L.J. Grimm, S. Ghate et al., Benefits and harms of breast cancer screening: a systematic review. JAMA 314(15), 1615–1634 (2015). https://doi.org/10.1001/jama.2015.13183
M. Román, M. Sala, L. Domingo, M. Posso, J. Louro, X. Castells, Personalized breast cancer screening strategies: a systematic review and quality assessment. PLoS One 14(12), e0226352 (2019). https://doi.org/10.1371/journal.pone.0226352
M. Escala-Garcia, A. Morra, S. Canisius, J. Chang-Claude, S. Kar, W. Zheng, et al., Breast cancer risk factors and their effects on survival: a Mendelian randomisation study. BMC Med. 18(1), 327 (2020). https://doi.org/10.1186/s12916-020-01797-2
M.R. Ataollahi, J. Sharifi, M.R. Paknahad, A. Paknahad, Breast cancer and associated factors: a review. J. Med. Life 8(Special Issue 4), 6–11 (2015)
Y.-S. Sun, Z. Zhao, Z.-N. Yang, F. Xu, H.-J. Lu, Z.-Y. Zhu et al., Risk factors and preventions of breast cancer. Int. J. Biol. Sci. 13(11), 1387–1397 (2017). https://doi.org/10.7150/ijbs.21635
S. Raichand, A.G. Dunn, M.-S. Ong, F.T. Bourgeois, E. Coiera, K.D. Mandl, Conclusions in systematic reviews of mammography for breast cancer screening and associations with review design and author characteristics. Syst. Rev. 6(1), 105 (2017). https://doi.org/10.1186/s13643-017-0495-6
M. Broeders, S. Moss, L. Nyström, S. Njor, H. Jonsson, E. Paap, et al., The impactof mammographic screening on breast cancer mortality in Europe: a review of observational studies. J. Med. Screen 19(Suppl 1), 14–25 (2012). https://doi.org/10.1258/jms.2012.012078
C. van den Ende, A.M. Oordt-Speets, H. Vroling, H.M.E. van Agt, Benefits and harms of breast cancer screening with mammography in women aged 40–49 years: a systematic review. Int. J. Cancer 141(7), 1295–1306 (2017). https://doi.org/10.1002/ijc.30794
H.G. Welch, P.C. Prorok, A.J. O’Malley, B.S. Kramer, Breast-cancer tumor size. Overdiag. Mammogr. Screen. Effect. 375(15), 1438–1447 (2016). https://doi.org/10.1056/NEJMoa1600249
J.S. Mandelblatt, N.K. Stout, C.B. Schechter, J.J. van den Broek, D.L. Miglioretti, M. Krapcho, et al., Collaborative modeling of the benefits and harms associated with different U.S. breast cancer screening strategies. Ann. Intern. Med. 164(4), 215–225 (2016). https://doi.org/10.7326/m15-1536
E. Warner, H. Messersmith, P. Causer, A. Eisen, R. Shumak, D. Plewes, Systematic review: using magnetic resonance imaging to screen women at high risk for breast cancer. Ann. Intern. Med. 148(9), 671–679 (2008). https://doi.org/10.7326/0003-4819-148-9-200805060-00007
S. Radhakrishna, S. Agarwal, P.M. Parikh, K. Kaur, S. Panwar, S. Sharma et al., Role of magnetic resonance imaging in breast cancer management. South Asian J Cancer 7(2), 69–71 (2018). https://doi.org/10.4103/sajc.sajc_104_18
L.W. Turnbull, Dynamic contrast-enhanced MRI in the diagnosis and management of breast cancer. NMR Biomed. 22(1), 28–39 (2009). https://doi.org/10.1002/nbm.1273
J. Xiao, H. Rahbar, D.S. Hippe, M.H. Rendi, E.U. Parker, N. Shekar, et al., Dynamic contrast-enhanced breast MRI features correlate with invasive breast cancer angiogenesis. npj Breast Cancer 7(1), 42 (2021). https://doi.org/10.1038/s41523-021-00247-3
N. Amornsiripanitch, S. Bickelhaupt, H.J. Shin, M. Dang, H. Rahbar, K. Pinker, et al., Diffusion-weighted MRI for unenhanced breast cancer screening, 293(3), 504–520 (2019). https://doi.org/10.1148/radiol.2019182789
S.C. Partridge, N. Amornsiripanitch, DWI in the assessment of breast lesions. Top. Magn. Reson. Imaging: TMRI 26(5), 201–209 (2017). https://doi.org/10.1097/RMR.0000000000000137
S.C. Partridge, E.S. McDonald, Diffusion weighted magnetic resonance imaging of the breast: protocol optimization, interpretation, and clinical applications. Magn. Reson. Imaging Clin. N. Am. 21(3), 601–624 (2013). https://doi.org/10.1016/j.mric.2013.04.007
J.K.P. Begley, T.W. Redpath, P.J. Bolan, F.J. Gilbert, In vivo proton magnetic resonance spectroscopy of breast cancer: a review of the literature. Breast Cancer Res. 14(2), 207 (2012). https://doi.org/10.1186/bcr3132
M.A. Bilal Ahmadani, S. Bhatty, Z.U. Abideen, M.S. Yaseen, T. Laique, J. Malik, Imaging in breast cancer: use of magnetic resonance spectroscopy.Cureus (2020). https://doi.org/10.7759/cureus.9734
W.A. Berg, Nuclear breast imaging: clinical results and future directions. J. Nucl. Med. 57(Supplement 1), 46S (2016). https://doi.org/10.2967/jnumed.115.157891
L.R. Greene, D. Wilkinson, The role of general nuclear medicine in breast cancer. J. Med. Radiat. Sci. 62(1), 54–65 (2015). https://doi.org/10.1002/jmrs.97
A. Daskin, A simple quantum neural net with a periodic activation function, in 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), 7–10 October 2018, pp. 2887–2891
N. Mishra, A. Bisarya, S. Kumar, B.K. Behera, S. Mukhopadhyay, P.K. Panigrahi, Cancer detection using quantum neural networks: a demonstration on a quantum computer (2019)
V. Azevedo, C. Silva, I. Dutra, Quantum transfer learning for breast cancerdetection. Quant. Mach. Intell. 4(1), 1–14 (2022b)
H.G. Kim, Y. Choi, Y.M. Ro, Modality-Bridge Transfer Learning for Medical Image Classification (IEEE, 2017), pp. 1–5
C.-K. Shie, C.-H. Chuang, C.-N. Chou, M.-H. Wu, E.Y. Chang, Transfer RepresentationLearning for Medical Image Analysis (IEEE, 2015), pp. 711–714
M.A. Morid, A. Borjali, G. Del Fiol, A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 128, 104115 (2021)
S. Lu, Z. Lu, Y.-D. Zhang, Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 30, 41–47 (2019). https://doi.org/10.1016/j.jocs.2018.11.008
S.-H. Wang, S. Xie, X. Chen, D.S. Guttery, C. Tang, J. Sun, et al., Alcoholism identification based on an AlexNet transfer learning model (original research), 10 (2019). https://doi.org/10.3389/fpsyt.2019.00205
P. Dauphin-Ducharme, K. Yang, N. Arroyo-Currás, K.L. Ploense, Y. Zhang, J. Gerson, et al., Electrochemical aptamer-based sensors for improved therapeutic drug monitoring and high-precision, feedback-controlled drug delivery. ACS Sens. 4(10), 2832–2837 (2019). https://doi.org/10.1021/acssensors.9b01616
E.L. Omonigho, M. David, A. Adejo, S. Aliyu, Breast cancer: tumor detectionin mammogram images using modified AlexNet deep convolution neural network, in 2020 International Conference in Mathematics, Computer Engineering and Computer Science (ICMCECS), 18–21 March 2020, pp. 1–6
F. Yan, X. Huang, Y. Yao, M. Lu, M. Li, Combining LSTM and DenseNet for automatic annotation and classification of chest x-ray images. IEEE Access 7, 74181–74189 (2019). https://doi.org/10.1109/ACCESS.2019.2920397
F. Nunnari, C. Bhuvaneshwara, A.O. Ezema, D. Sonntag, A study on the fusion of pixels and patient metadata in CNN-based classification of skin lesion images, in Machine Learning and Knowledge Extraction, ed. by A. Holzinger, P. Kieseberg, A.M. Tjoa, E. Weippl (Springer International Publishing, Cham, 2020), pp. 191–208
P.U. Hepsağ, S.A. Özel, A. Yazıcı, Using deep learning for mammography classification, 418–423 (2017)
J. Diz, G. Marreiros, A. Freitas, Applying data mining techniques to improve breast cancer diagnosis. J. Med. Syst. 40(9), 203 (2016). https://doi.org/10.1007/s10916-016-0561-y
V. Azevedo, C. Silva, I. Dutra, Quantum transfer learning for breast cancer detection. Quant. Mach. Intell. 4(1), 5 (2022a). https://doi.org/10.1007/s42484-022-00062-4
A. Ahuja, L. Al-Zogbi, A. Krieger, Application of noise-reduction techniques tomachine learning algorithms for breast cancer tumor identification. Comput. Biol. Med. 135, 104576 (2021). https://doi.org/10.1016/j.compbiomed.2021.104576
K. Gupta, R.R. Janghel, Dimensionality reduction-based breast cancer classification using machine learning, in Computational Intelligence: Theories, Applications and Future Directions—Volume I, ed. by N.K. Verma, & A.K. Ghosh (Springer, Singapore, 2019), pp. 133–146
D.A. Omondiagbe, S. Veeramani, A.S. Sidhu, Machine learning classification techniques for breast cancer diagnosis. IOP Conference Series: Materials Science and Engineering 495, 012033 (2019). https://doi.org/10.1088/1757-899x/495/1/012033
M.F. Akay, Support vector machines combined with feature selection for breast cancer diagnosis. Exp. Syst. Appl. 36(2, Part 2), 3240–3247 (2009). https://doi.org/10.1016/j.eswa.2008.01.009
A.T. Azar, S.A. El-Said, Performance analysis of support vector machines classifiers in breast cancer mammography recognition. Neural Comput. Appl. 24(5), 1163–1177 (2014). https://doi.org/10.1007/s00521-012-1324-4
B.M. Salih Hasan, A.M. Abdulazeez, A review of principal component analysis algorithm for dimensionality reduction. J. Soft Comput. Data Min. 2(1), 20–30. https://publisher.uthm.edu.my/ojs/index.php/jscdm/article/view/8032
M.D. Ritchie, L.W. Hahn, N. Roodi, L.R. Bailey, W.D. Dupont, F.F. Parl et al., Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 69(1), 138–147 (2001). https://doi.org/10.1086/321276
X. Liu, J. Shi, S. Zhou, M. Lu, An iterated Laplacian based semi-superviseddimensionality reduction for classification of breast cancer on ultrasound images, in 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 26–30 August 2014, pp. 4679–4682
C. Deisy, B. Subbulakshmi, S. Baskar, N Ramaraj, Efficient dimensionalityreduction approaches for feature selection, in International Conference on Computational Intelligence and Multimedia Applications (ICCIMA 2007), 13–15 December 2007, pp. 121–127
A. Jamal, A. Handayani, A.A. Septiandri, E. Ripmiatin, Y. Effendi, Dimensionality reduction using PCA and k-means clustering for breast cancer prediction. Lontar Komput. J. Ilm. Teknol. Inf 9(3), 192–201 (2018)
W.S. Noble, What is a support vector machine? Nat. Biotechnol. 24(12), 1565–1567 (2006)
S. Suthaharan, Support vector machine, in Machine Learning Models and Algorithms for Big Data Classification (Springer, 2016), pp. 207–235
A. Widodo, B.-S. Yang, Support vector machine in machine condition monitoring and fault diagnosis, 21(6), 2560–2574 (2007)
D.A. Pisner, D.M. Schnyer, Support Vector Machine. Machine Learning. (Elsevier, 2020), pp. 101–121
T. Joachims, SVM light is an implementation of support vector machines (SVMs), in C. University of Dortmund, Collaborative Research Center on Complexity Reduction in Multivariate Data (SFB475); gmd.de/∼thorsten/svm_light (2000)
H.X. Liu, R.S. Zhang, F. Luan, X.J. Yao, M.C. Liu, Z.D. Hu et al., Diagnosing breast cancer based on support vector machines. J. Chem. Inf. Comput. Sci. 43(3), 900–907 (2003). https://doi.org/10.1021/ci0256438
M.E. Ozer, P.O. Sarica, K.Y. Arga, New machine learning applications to accelerate personalized medicine in breast cancer: rise of the support vector machines. Omics: J. Integrative Biol. 24(5), 241–246 (2020)
S. Ghosh,S. Mondal, B. Ghosh, A Comparative Study of Breast Cancer Detection Based on SVM and MLP BPN Classifier (IEEE, 2014), pp. 1–4
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Prajapati, J.B., Paliwal, H., Prajapati, B.G., Saikia, S., Pandey, R. (2023). Quantum Machine Learning in Prediction of Breast Cancer. In: Pandey, R., Srivastava, N., Singh, N.K., Tyagi, K. (eds) Quantum Computing: A Shift from Bits to Qubits. Studies in Computational Intelligence, vol 1085. Springer, Singapore. https://doi.org/10.1007/978-981-19-9530-9_19
Download citation
DOI: https://doi.org/10.1007/978-981-19-9530-9_19
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-9529-3
Online ISBN: 978-981-19-9530-9
eBook Packages: EngineeringEngineering (R0)