Abstract
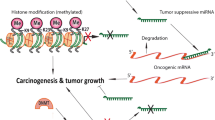
Recent studies have shown that the alterations in the expression of miRNAs play critical roles in the processes of carcinogenesis and cancer progression. During the process of carcinogenesis, gene mutation and epigenetic aberration together with the exposure of environmental factors such as infection, ultraviolet light, irradiation, and environmental toxicants could change the expression level of miRNAs, resulting in the alterations in gene expression and cellular signal transductions. The aberrant expression of miRNAs initiates carcinogenesis and subsequently promotes the progression of cancers. Therefore, targeting the altered miRNAs may be a new approach for cancer prevention and treatment. Interestingly, dietary agents (natural agents) collectively known as nutraceuticals have been found to modulate and normalize the expression of miRNAs. Thus, the non-toxic dietary natural agents such as isoflavone genistein, curcumin, resveratrol, I3C, DIM, vitamins, etc. may have effects on cancer prevention and treatment. The in vitro and in vivo studies have provided the evidences showing that these agents could up-regulate tumor suppressive miRNAs and down-regulate oncogenic miRNAs, resulting in the suppression of cancer formation and progression. Hence, these agents could prevent the occurrence of cancer and may also inhibit tumor progression. Moreover, treatment with these non-toxic agents together with chemotherapeutics may be a novel approach for cancer therapy.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Nair S, Li W, Kong AN. Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharmacol Sin. 2007;28:459–72.
Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610.
Zhao L, Bode AM, Cao Y, et al. Regulatory mechanisms and clinical perspectives of miRNA in tumor radiosensitivity. Carcinogenesis. 2012;33:2220–7.
Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34.
Manikandan J, Aarthi JJ, Kumar SD, et al. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2:330–4.
Lee YS, Kim HK, Chung S, et al. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–41.
Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Busbee PB, Nagarkatti M, Nagarkatti PS. Natural indoles, indole-3-carbinol and 3,3′-diindolymethane, inhibit T cell activation by staphylococcal enterotoxin B through epigenetic regulation involving HDAC expression. Toxicol Appl Pharmacol. 2014;274:7–16.
Hegarty M, Coate J, Sherman-Broyles S, et al. Lessons from natural and artificial polyploids in higher plants. Cytogenet Genome Res. 2013;140:204–25.
Li Y, Chen H, Hardy TM, et al. Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS One. 2013;8:e54369.
Li Y, Kong D, Wang Z, et al. Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res. 2010;27:1027–41.
Li Y, Kong D, Ahmad A, et al. Epigenetic deregulation of miR-29a and miR-1256 by isoflavone contributes to the inhibition of prostate cancer cell growth and invasion. Epigenetics. 2012;7:940–9.
Sun G, Yan J, Noltner K, et al. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–51.
Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7.
Yu Z, Li Z, Jolicoeur N, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–41.
Blitzblau RC, Weidhaas JB. MicroRNA binding-site polymorphisms as potential biomarkers of cancer risk. Mol Diagn Ther. 2010;14:335–42.
Pelletier C, Weidhaas JB. MicroRNA binding site polymorphisms as biomarkers of cancer risk. Expert Rev Mol Diagn. 2010;10:817–29.
Akkiz H, Bayram S, Bekar A, et al. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat. 2011;18:e399–407.
Chu YH, Tzeng SL, Lin CW, et al. Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLoS One. 2012;7:e39777.
Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40.
Zhang W, Winder T, Ning Y, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–9.
Izzotti A, Larghero P, Longobardi M, et al. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat Res. 2011;717:9–16.
Izzotti A, Calin GA, Steele VE, et al. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila). 2010;3:62–72.
Izzotti A, Calin GA, Arrigo P, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–12.
Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–4.
Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004.
Izzotti A. Molecular medicine and the development of cancer chemopreventive agents. Ann N Y Acad Sci. 2012;1259:26–32.
Izzotti A, Larghero P, Cartiglia C, et al. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010;31:894–901.
Schembri F, Sridhar S, Perdomo C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:2319–24.
Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–41.
Russ R, Slack FJ. Cigarette-smoke-induced dysregulation of microRNA expression and its role in lung carcinogenesis. Pulm Med. 2012;2012:791234.
Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science. 2014;346:945–9.
Syed DN, Khan MI, Shabbir M, et al. MicroRNAs in skin response to UV radiation. Curr Drug Targets. 2013;14:1128–34.
Li W, Zhou BR, Hua LJ, et al. Differential miRNA profile on photoaged primary human fibroblasts irradiated with ultraviolet A. Tumour Biol. 2013;34:3491–500.
Kraemer A, Chen IP, Henning S, et al. UVA and UVB irradiation differentially regulate microRNA expression in human primary keratinocytes. PLoS One. 2013;8:e83392.
Greenberg E, Rechavi G, Amariglio N, et al. Mutagen-specific mutation signature determines global microRNA binding. PLoS One. 2011;6:e27400.
Lal A, Pan Y, Navarro F, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–8.
Brunner S, Herndler-Brandstetter D, Arnold CR, et al. Upregulation of miR-24 is associated with a decreased DNA damage response upon etoposide treatment in highly differentiated CD8(+) T cells sensitizing them to apoptotic cell death. Aging Cell. 2012;11:579–87.
Wang XC, Du LQ, Tian LL, et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer. 2011;72:92–9.
Zhang Y, Wei W, Cheng N, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631–40.
Honegger A, Schilling D, Bastian S, et al. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015;11:e1004712.
Paiva I, Gil da Costa RM, Ribeiro J, et al. A role for MicroRNA-155 expression in microenvironment associated to HPV-induced carcinogenesis in K14-HPV16 transgenic mice. PLoS One. 2015;10:e0116868.
Miller DL, Davis JW, Taylor KH, et al. Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the cancer genome atlas. Am J Pathol. 2015;185:679–92.
Shishodia G, Verma G, Srivastava Y, et al. Deregulation of microRNAs Let-7a and miR-21 mediate aberrant STAT3 signaling during human papillomavirus-induced cervical carcinogenesis: role of E6 oncoprotein. BMC Cancer. 2014;14:996.
Wang X, Wang HK, McCoy JP, et al. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–47.
Yu H, Lu J, Zuo L, et al. Epstein-Barr virus downregulates microRNA 203 through the oncoprotein latent membrane protein 1: a contribution to increased tumor incidence in epithelial cells. J Virol. 2012;86:3088–99.
Xu C, Zheng Y, Lian D, et al. Analysis of MicroRNA expression profile identifies novel biomarkers for non-small cell lung cancer. Tumori. 2015;101:104–10.
Huang P, Ye B, Yang Y, et al. MicroRNA-181 functions as a tumor suppressor in non-small cell lung cancer (NSCLC) by targeting Bcl-2. Tumour Biol. 2014;36:3381–7.
Geng Q, Fan T, Zhang B, et al. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149.
Zhu W, He J, Chen D, et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;9:e87780.
Nadal E, Zhong J, Lin J, et al. A MicroRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res. 2014;20:3107–17.
Drusco A, Nuovo GJ, Zanesi N, et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One. 2014;9:e96670.
Hofsli E, Sjursen W, Prestvik WS, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108:1712–9.
Christensen LL, Tobiasen H, Holm A, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer. 2013;133:67–78.
He X, Dong Y, Wu CW, et al. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol Med. 2012;18:1491–8.
Sun JY, Huang Y, Li JP, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun. 2012;420:787–92.
Zhang Y, He X, Liu Y, et al. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep. 2012;27:685–94.
Song C, Chen H, Wang T, et al. Expression profile analysis of microRNAs in prostate cancer by next-generation sequencing. Prostate. 2015;75:500–16.
Hart M, Nolte E, Wach S, et al. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol Cancer Res. 2014;12:250–63.
Reis ST, Pontes-Junior J, Antunes AA, et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012;12:14.
Rane JK, Scaravilli M, Ylipaa A, et al. MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur Urol. 2015;67:7–10.
Kong D, Heath E, Chen W, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am J Transl Res. 2012;4:14–23.
Zearo S, Kim E, Zhu Y, et al. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer. 2014;14:200.
Cui W, Zhang S, Shan C, et al. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962–74.
Okuda H, Xing F, Pandey PR, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–44.
Wang B, Wang H, Yang Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS One. 2012;7:e47053.
Sun X, Luo S, He Y, et al. Screening of the miRNAs related to breast cancer and identification of its target genes. Eur J Gynaecol Oncol. 2014;35:696–700.
Erturk E, Cecener G, Tezcan G, et al. BRCA mutations cause reduction in miR-200c expression in triple negative breast cancer. Gene. 2015;556:163–9.
Danza K, De SS, Pinto R, et al. MiR-578 and miR-573 as potential players in BRCA-related breast cancer angiogenesis. Oncotarget. 2015;6:471–83.
Li P, Sheng C, Huang L, et al. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014;16:473.
Su A, He S, Tian B, et al. MicroRNA-221 mediates the effects of PDGF-BB on migration, proliferation, and the epithelial-mesenchymal transition in pancreatic cancer cells. PLoS One. 2013;8:e71309.
Li CH, To KF, Tong JH, et al. Enhancer of zeste homolog 2 silences microRNA-218 in human pancreatic ductal adenocarcinoma cells by inducing formation of heterochromatin. Gastroenterology. 2013;144:1086–97.
Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol. 2012;25:1609–22.
Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71.
Shi S, Lu Y, Qin Y, et al. miR-1247 is correlated with prognosis of pancreatic cancer and inhibits cell proliferation by targeting neuropilins. Curr Mol Med. 2014;14:316–27.
Arora S, Swaminathan SK, Kirtane A, et al. Synthesis, characterization, and evaluation of poly (D, L-lactide-co-glycolide)-based nanoformulation of miRNA-150: potential implications for pancreatic cancer therapy. Int J Nanomedicine. 2014;9:2933–42.
Wei L, Lian B, Zhang Y, et al. Application of microRNA and mRNA expression profiling on prognostic biomarker discovery for hepatocellular carcinoma. BMC Genomics. 2014;15 Suppl 1:S13.
Koufaris C, Wright J, Currie RA, et al. Hepatic microRNA profiles offer predictive and mechanistic insights after exposure to genotoxic and epigenetic hepatocarcinogens. Toxicol Sci. 2012;128:532–43.
Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–32.
Wong QW, Ching AK, Chan AW, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–75.
Zheng F, Liao YJ, Cai MY, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–89.
Shinozaki-Ushiku A, Kunita A, Isogai M, et al. Profiling of virus-encoded microRNAs in Epstein-Barr virus-associated gastric carcinoma and their roles in gastric carcinogenesis. J Virol. 2015;89:5581–91.
Sun J, Song Y, Wang Z, et al. Clinical significance of promoter region hypermethylation of microRNA-148a in gastrointestinal cancers. Onco Targets Ther. 2014;7:853–63.
Li Z, Lei H, Luo M, et al. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54.
Amodio N, Di Martino MT, Foresta U, et al. MiR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436.
Amodio N, Rossi M, Raimondi L, et al. miR-29 s: a family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget. 2015;6:12837–61.
Yang Y, Li F, Saha MN, et al. miR-137 and miR-197 Induce Apoptosis and Suppress Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin Cancer Res. 2015;21:2399–411.
Zhao JJ, Lin J, Zhu D, et al. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/beta-catenin/BCL9 pathway. Cancer Res. 2014;74:1801–13.
Peng J, Thakur A, Zhang S, et al. Expressions of miR-181a and miR-20a in RPMI8226 cell line and their potential as biomarkers for multiple myeloma. Tumour Biol. 2015;36:8545–52.
Du J, Liu S, He J, et al. MicroRNA-451 regulates stemness of side population cells via PI3K/Akt/mTOR signaling pathway in multiple myeloma. Oncotarget. 2015;6:14993–5007.
Rossi M, Amodio N, Di Martino MT, et al. MicroRNA and multiple myeloma: from laboratory findings to translational therapeutic approaches. Curr Pharm Biotechnol. 2014;15:459–67.
Amodio N, Di Martino MT, Neri A, et al. Non-coding RNA: a novel opportunity for the personalized treatment of multiple myeloma. Expert Opin Biol Ther. 2013;13 Suppl 1:S125–37.
Rossi M, Amodio N, Di Martino MT, et al. From target therapy to miRNA therapeutics of human multiple myeloma: theoretical and technological issues in the evolving scenario. Curr Drug Targets. 2013;14:1144–9.
Soga D, Yoshiba S, Shiogama S, et al. microRNA expression profiles in oral squamous cell carcinoma. Oncol Rep. 2013;30:579–83.
Ko MA, Zehong G, Virtanen C, et al. MicroRNA expression profiling of esophageal cancer before and after induction chemoradiotherapy. Ann Thorac Surg. 2012;94:1094–102.
Weber F, Teresi RE, Broelsch CE, et al. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91:3584–91.
Andrade TA, Evangelista AF, Campos AH, et al. A microRNA signature profile in EBV+ diffuse large B-cell lymphoma of the elderly. Oncotarget. 2014;5:11813–26.
Sandoval J, Diaz-Lagares A, Salgado R, et al. MicroRNA expression profiling and DNA methylation signature for deregulated microRNA in cutaneous T-cell lymphoma. J Invest Dermatol. 2014;135:1128–37.
Koens L, Qin Y, Leung WY, et al. MicroRNA profiling of primary cutaneous large B-cell lymphomas. PLoS One. 2013;8:e82471.
Motsch N, Alles J, Imig J, et al. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS One. 2012;7:e42193.
Schotte D, De Menezes RX, Akbari MF, et al. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96:703–11.
Robertus JL, Kluiver J, Weggemans C, et al. MiRNA profiling in B non-Hodgkin lymphoma: a MYC-related miRNA profile characterizes Burkitt lymphoma. Br J Haematol. 2010;149:896–9.
Gibcus JH, Tan LP, Harms G, et al. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–76.
Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–40.
Neelakandan K, Babu P, Nair S. Emerging roles for modulation of microRNA signatures in cancer chemoprevention. Curr Cancer Drug Targets. 2012;12:716–40.
Wang Y, Li Y, Liu X, et al. Genetic and epigenetic studies for determining molecular targets of natural product anticancer agents. Curr Cancer Drug Targets. 2013;13:506–18.
Messina MJ. Legumes and soybeans: overview of their nutritional profiles and health effects. Am J Clin Nutr. 1999;70:439S–50S.
Messina MJ, Persky V, Setchell KD, et al. Soy intake and cancer risk: a review of the in vitro and in vivo data. Nutr Cancer. 1994;21:113–31.
Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3:503–18.
Zhang Y, Chen H. Genistein, an epigenome modifier during cancer prevention. Epigenetics. 2011;6:888–91.
Rabiau N, Trraf HK, Adjakly M, et al. miRNAs differentially expressed in prostate cancer cell lines after soy treatment. In Vivo. 2011;25:917–21.
Hirata H, Hinoda Y, Shahryari V, et al. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer. 2014;110:1645–54.
Chen Y, Zaman MS, Deng G, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila). 2011;4:76–86.
Zaman MS, Chen Y, Deng G, et al. The functional significance of microRNA-145 in prostate cancer. Br J Cancer. 2010;103:256–64.
Xia J, Cheng L, Mei C, et al. Genistein inhibits cell growth and invasion through regulation of miR-27a in pancreatic cancer cells. Curr Pharm Des. 2014;20:5348–53.
Sun Q, Cong R, Yan H, et al. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol Rep. 2009;22:563–7.
Li Y, Vandenboom TG, Wang Z, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–95.
Li Y, Vandenboom TG, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–12.
Parker LP, Taylor DD, Kesterson J, et al. Modulation of microRNA associated with ovarian cancer cells by genistein. Eur J Gynaecol Oncol. 2009;30:616–21.
Recio MC, Andujar I, Rios JL. Anti-inflammatory agents from plants: progress and potential. Curr Med Chem. 2012;19:2088–103.
Krishnaswamy K. Traditional Indian spices and their health significance. Asia Pac J Clin Nutr. 2008;17 Suppl 1:265–8.
Prasad S, Gupta SC, Tyagi AK, et al. Curcumin, a component of golden spice: from bedside to bench and back. Biotechnol Adv. 2014;32:1053–64.
Park W, Amin AR, Chen ZG, et al. New perspectives of curcumin in cancer prevention. Cancer Prev Res (Phila). 2013;6:387–400.
Saini S, Arora S, Majid S, et al. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res (Phila). 2011;4:1698–709.
Teiten MH, Dicato M, Diederich M. Curcumin as a regulator of epigenetic events. Mol Nutr Food Res. 2013;57:1619–29.
Fu S, Kurzrock R. Development of curcumin as an epigenetic agent. Cancer. 2010;116:4670–6.
Zheng J, Wu C, Lin Z, et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation—a novel mechanism suppressing liver fibrosis. FEBS J. 2014;281:88–103.
Kong LM, Liao CG, Zhang Y, et al. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014;74:3764–78.
Yang J, Cao Y, Sun J, et al. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med Oncol. 2010;27:1114–8.
Zhang J, Du Y, Wu C, et al. Curcumin promotes apoptosis in human lung adenocarcinoma cells through miR-186* signaling pathway. Oncol Rep. 2010;24:1217–23.
Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol. 2010;73:185–91.
Roy S, Levi E, Majumdar AP, et al. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58.
Bao B, Ali S, Banerjee S, et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335–45.
Giovinazzo G, Ingrosso I, Paradiso A, et al. Resveratrol biosynthesis: plant metabolic engineering for nutritional improvement of food. Plant Foods Hum Nutr. 2012;67:191–9.
Shukla Y, Singh R. Resveratrol and cellular mechanisms of cancer prevention. Ann N Y Acad Sci. 2011;1215:1–8.
Jha RK, Ma Q, Sha H, et al. Emerging role of resveratrol in the treatment of severe acute pancreatitis. Front Biosci (Schol Ed). 2010;2:168–75.
Aluyen JK, Ton QN, Tran T, et al. Resveratrol: potential as anticancer agent. J Diet Suppl. 2012;9:45–56.
Qin W, Zhang K, Clarke K, et al. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutr Cancer. 2014;66:270–7.
Yu YH, Chen HA, Chen PS, et al. MiR-520 h-mediated FOXC2 regulation is critical for inhibition of lung cancer progression by resveratrol. Oncogene. 2013;32:431–43.
Dhar S, Hicks C, Levenson AS. Resveratrol and prostate cancer: promising role for microRNAs. Mol Nutr Food Res. 2011;55:1219–29.
Cao Z, Yoon JH, Nam SW, et al. PDCD4 expression inversely correlated with miR-21 levels in gastric cancers. J Cancer Res Clin Oncol. 2012;138:611–9.
Sheth S, Jajoo S, Kaur T, et al. Resveratrol reduces prostate cancer growth and metastasis by inhibiting the Akt/MicroRNA-21 pathway. PLoS One. 2012;7:e51655.
Tili E, Michaille JJ, Alder H, et al. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGFbeta signaling pathway in SW480 cells. Biochem Pharmacol. 2010;80:2057–65.
Han Z, Yang Q, Liu B, et al. MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis. 2012;33:131–9.
Bae S, Lee EM, Cha HJ, et al. Resveratrol alters microRNA expression profiles in A549 human non-small cell lung cancer cells. Mol Cells. 2011;32:243–9.
Minich DM, Bland JS. A review of the clinical efficacy and safety of cruciferous vegetable phytochemicals. Nutr Rev. 2007;65:259–67.
Higdon JV, Delage B, Williams DE, et al. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55:224–36.
Acharya A, Das I, Singh S, et al. Chemopreventive properties of indole-3-carbinol, diindolylmethane and other constituents of cardamom against carcinogenesis. Recent Pat Food Nutr Agric. 2010;2:166–77.
Firestone GL, Sundar SN. Minireview: modulation of hormone receptor signaling by dietary anticancer indoles. Mol Endocrinol. 2009;23:1940–7.
Firestone GL, Bjeldanes LF. Indole-3-carbinol and 3-3′-diindolylmethane antiproliferative signaling pathways control cell-cycle gene transcription in human breast cancer cells by regulating promoter-Sp1 transcription factor interactions. J Nutr. 2003;133:2448S–55.
Melkamu T, Zhang X, Tan J, et al. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31:252–8.
Mao HL, Zhu ZQ, Chen CD. The androgen receptor in hormone-refractory prostate cancer. Asian J Androl. 2009;11:69–73.
Rossi A, D’Urso OF, Gatto G, et al. Non-coding RNAs change their expression profile after Retinoid induced differentiation of the promyelocytic cell line NB4. BMC Res Notes. 2010;3:24.
Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–57.
Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–8.
Wang LL, Zhang Z, Li Q, et al. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod. 2009;24:562–79.
Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–8.
Wang X, Gocek E, Liu CG, et al. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8:736–41.
Alvarez-Diaz S, Valle N, Ferrer-Mayorga G, et al. MicroRNA-22 is induced by vitamin D and contributes to its antiproliferative, antimigratory and gene regulatory effects in colon cancer cells. Hum Mol Genet. 2012;21:2157–65.
Mohri T, Nakajima M, Takagi S, et al. MicroRNA regulates human vitamin D receptor. Int J Cancer. 2009;125:1328–33.
Gaedicke S, Zhang X, Schmelzer C, et al. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett. 2008;582:3542–6.
Sarveswaran S, Liroff J, Zhou Z, et al. Selenite triggers rapid transcriptional activation of p53, and p53-mediated apoptosis in prostate cancer cells: Implication for the treatment of early-stage prostate cancer. Int J Oncol. 2010;36:1419–28.
Acknowledgements
The authors’ work cited in this review article was funded by grants from the National Cancer Institute, NIH (R01CA108535, R01CA154321, and R01CA164318 awarded to FHS). We also thank Puschelberg and Guido foundations for their generous contribution.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Li, Y., Sarkar, F.H. (2016). Targeting MicroRNAs: Molecular Basis of Cancer Prevention. In: Chatterjee, M. (eds) Molecular Targets and Strategies in Cancer Prevention. Springer, Cham. https://doi.org/10.1007/978-3-319-31254-5_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-31254-5_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-31252-1
Online ISBN: 978-3-319-31254-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)