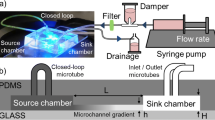
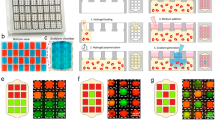
Abstract
Biomolecular gradients are widely present in multiple biological processes. Historically they were reproduced in vitro by using micropipettes, Boyden and Zigmond chambers, or hydrogels. Despite the great utility of these setups in the study of gradient-related problems such as chemotaxis, they face limitations when trying to translate more complex in vivo-like scenarios to in vitro systems. In the last 20 years, the advances in manufacturing of micromechanical systems (MEMS) had opened the possibility of applying this technology to biology (BioMEMS). In particular, microfluidics has proven extremely efficient in setting-up biomolecular gradients which are stable, controllable, reproducible and at length scales that are relevant to cells. In this chapter, we give an overview of different methods to generate molecular gradients using microfluidics, then we discuss the different steps of the pipeline to fabricate a gradient generator microfluidic device, and at the end, we show an application example of the fabrication of a microfluidic device that can be used to generate a surface-bound biomolecular gradient.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Pearsall J et al (2012) Oxford dictionaries online. Oxford Univ. Press
Bogenrieder T, Herlyn M (2003) Axis of evil: molecular mechanisms of cancer metastasis. Oncogene 22:6524–6536
Wang S-J et al (2004) Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res 300:180–189
Janeway CA, Travers P, Walport M, Shlomchik MJ (2001) Immunobiology: the immune system in health and disease, 5th edn. Garland
Goodman CS (1996) Mechanisms and molecules that control growth cone guidance. Annu Rev Neurosci 19:341–377
Stossel TP (1993) On the crawling of animal cells. Science 260:1086–1094
Redd MJ, Kelly G, Dunn G, Way M, Martin P (2006) Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation. Cell Motil Cytoskeleton 63:415–422
Chaudhury MK, Whitesides GM (1992) How to make water run uphill. Science 256:1539–1541
Morgenthaler S, Lee S, Zürcher S, Spencer ND, Zu S, Simple A (2003) Reproducible approach to the preparation of surface-chemical gradients. Langmuir 19:988–989
Jeon NL et al (2000) Generation of solution and surface gradients using microfluidic systems. Langmuir 16:8311–8316
Genzer J, Bhat RR (2008) Surface-bound soft matter gradients. Langmuir 24:2294–2317
Meredith JC, Karim A, Amis EJ (2002) Combinatorial methods for investigations in polymer materials science. MRS 27:330–335
Grayson ACR et al (2004) A BioMEMS review: MEMS technology for physiologically integrated devices. Proc IEEE 92:6–21
Trichet L et al (2012) Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci 109:6933–6938
Lin B, Hui J, Mao H (2021) Nanopore technology and its applications in gene sequencing. Biosensors 11:214
Heller I, Hoekstra TP, King GA, Peterman EJG, Wuite GJL (2014) Optical tweezers analysis of DNA-protein complexes. Chem Rev 114:3087–3119
Rigat-Brugarolas LG et al (2014) A functional microengineered model of the human splenon-on-a-chip. Lab Chip 14:1715–1724
Castillo-Fernandez O, Rodriguez-Trujillo R, Gomila G, Samitier J (2014) High-speed counting and sizing of cells in an impedance flow microcytometer with compact electronic instrumentation. Microfluid Nanofluidics 16:91–99
Holden MA, Kumar S, Castellana ET, Beskok A, Cremer PS (2003) Generating fixed concentration arrays in a microfluidic device. Sensors Actuators B Chem 92:199–207
Georgescu W et al (2008) Model-controlled hydrodynamic focusing to generate multiple overlapping gradients of surface-immobilized proteins in microfluidic devices. Lab Chip 8:238–244
Comelles J, Hortigüela V, Samitier J, Martínez E (2012) Versatile gradients of covalently bound proteins on microstructured substrates. Langmuir 28:13688–13697
Keenan TM, Hsu C-H, Folch A (2006) Microfluidic “jets” for generating steady-state gradients of soluble molecules on open surfaces. Appl Phys Lett 89:114103
Cosson S, Aeberli LG, Brandenberg N, Lutolf MP (2015) Ultra-rapid prototyping of flexible, multi-layered microfluidic devices via razor writing. Lab Chip 15:72–76
Duffy DC, McDonald JC, Schueller OJA, Whitesides GM (1998) Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal Chem 70:4974–4984
Ibidi (2021) μ-Slide chemotaxis
Comelles J, Hortigüela V, Martínez E, Riveline D (2015) Methods for rectifying cell motions in vitro: breaking symmetry using microfabrication and microfluidics. In: Paluch E (ed) Biophysical methods in cell biology. Elsevier Inc., pp 437–452
Hollingsworth K (2020) Chips and tips. Solvent extraction of 3D printed molds for soft lithography. Royal Society Chemistry
Acknowledgments
We thank the Martinez Lab members for their discussions and help. Funding was provided by a European Union Horizon 2020 ERC grant (agreement no. 647863—COMIET), ‘La Caixa’ Foundation (grant no. LCF/PR/HR20/52400011), the CERCA Programme/Generalitat de Catalunya (2017-SGR-1079), and the Spanish Ministry of Economy and Competitiveness (Severo Ochoa Program for Centers of Excellence in R&D 2016-2019). The results presented here only reflect the views of the authors; the European Commission is not responsible for any use that may be made of the information it contains.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Comelles, J., Castillo-Fernández, Ó., Martínez, E. (2022). How to Get Away with Gradients. In: Caballero, D., Kundu, S.C., Reis, R.L. (eds) Microfluidics and Biosensors in Cancer Research. Advances in Experimental Medicine and Biology, vol 1379. Springer, Cham. https://doi.org/10.1007/978-3-031-04039-9_2
Download citation
DOI: https://doi.org/10.1007/978-3-031-04039-9_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-04038-2
Online ISBN: 978-3-031-04039-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)