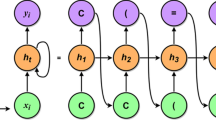
Abstract
Two generative autoencoder models for designing novel drug-like compounds able to block the catalytic site of the SARS-CoV-2 main protease (MPro) critical for mediating viral replication and transcription were developed using deep learning methods. To do this, the following steps were performed: (i) architectures of two neural networks were constructed; (ii) a virtual compound library of potential anti-SARS-CoV-2 MPro agents for training two neural networks was formed; (iii) molecular docking of all compounds from this library with MPro was made and calculations of the values of binding free energy were carried out; (iv) two neural networks were trained followed by estimation of the learning outcomes and work of two autoencoders involving several generation modes. Validation of autoencoders and their comparison revealed the best combination of the neural network architecture with the generation mode, which allows one to generate good chemical scaffold for the design of novel antiviral drugs with suitable pharmaceutical properties.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Iqbal, T., Qureshi, S.: The survey: text generation models in deep learning. J. King Saud Univ. Comput. Inf. Sci. (2020). https://doi.org/10.1016/j.jksuci.2020.04.001
Sorin, V., Barash, Y., Konen, E., Klang, E.: Creating artificial images for radiology applications using generative adversarial networks (GANs) – A systematic review. Acad. Radiol. 27(8), 1175–1185 (2020). https://doi.org/10.1016/j.acra.2019.12.024
Kim, S., et al.: PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 49(D1), D1388–D1395 (2021). https://doi.org/10.1093/nar/gkaa971
PubChem Homepage. https://pubchem.ncbi.nlm.nih.gov/. Accessed 10 Dec 2021
Chen, Y., Liu, Q., Guo, D.: Coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 92(4), 418–423 (2020). https://doi.org/10.1002/jmv.25681
Anand, K., Palm, G.J., Mesters, J.R., Siddell, S.G., Ziebuhr, J., Hilgenfeld, R.: Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 21(13), 3213–3224 (2002). https://doi.org/10.1093/emboj/cdf327
Yang, H., et al.: The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. PNAS USA 100(23), 13190–13195 (2003). https://doi.org/10.1073/pnas.1835675100
Hegyi, A., Ziebuhr, J.: Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 83(3), 595–599 (2002). https://doi.org/10.1099/0022-1317-83-3-595
Pillaiyar, T., Manickam, M., Namasivayam, V., Hayashi, Y., Jung, S.H.: An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 59(14), 6595–6628 (2016). https://doi.org/10.1021/acs.jmedchem.5b01461
Yan, F., Gao, F.: An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2. Comp. Struct. Biotechnol. J. 19, 4868–4883 (2021). https://doi.org/10.1016/j.csbj.2021.08.036
Ullrich, S., Nitsche, C.: The SARS-CoV-2 main protease as drug target. Bioorganic Med. Chem. Lett. 30(17), 127377 (2020). https://doi.org/10.1016/j.bmcl.2020.127377
Forster, P., Forster, L., Renfrew, C., Forster, M.: Phylogenetic network analysis of SARS-CoV-2 genomes. PNAS USA 117(17), 9241–9243 (2020). https://doi.org/10.1073/pnas.2004999117
Pachetti, M., et al.: Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 18, 179 (2020). https://doi.org/10.1186/s12967-020-02344-6
Yao, H., et al.: Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov. 6, 76 (2020). https://doi.org/10.1038/s41421-020-00226-1
Khailany, R.A., Safdar, M., Ozaslan, M.: Genomic characterization of a novel SARS-CoV-2. Gene Rep. 19, 100682 (2020). https://doi.org/10.1016/j.genrep.2020.100682
Weininger, D.: SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 28(1), 31–36 (1988). https://doi.org/10.1021/ci00057a005
Andrianov, A.M., Nikolaev, G.I., Shuldov, N.A., Bosko, I.P., Anischenko, A.I., Tuzikov, A.V.: Application of deep learning and molecular modeling to identify small drug-like compounds as potential HIV-1 entry inhibitors. J. Biomol. Struct. Dyn. 1–19 (2021). https://doi.org/10.1080/07391102.2021.1905559
Pharmit Homepage. http://pharmit.csb.pitt.edu. Accessed 10 Dec 2021
Sunseri, J., Koes, D.R.: Pharmit: interactive exploration of chemical space. Nucleic Acids Res. 44(W1), W442–W448 (2016). https://doi.org/10.1093/nar/gkw287
Schneidman-Duhovny, D., Dror, O., Inbar, Y., Nussinov, R., Wolfson, H.J.: Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules. J. Comput. Biol. 15(7), 737–754 (2008). https://doi.org/10.1089/cmb.2007.0130
PubChemPy Homepage. https://pubchempy.readthedocs.io/. Accessed 10 Dec 2021
Python Homepage. https://www.python.org/. Accessed 10 Dec 2021
RCSB PDB Homepage. https://www.rcsb.org/pdb/. Accessed 10 Dec 2021
RDKit Homepage. http://www.rdkit.org/. Accessed 10 Dec 2021
Halgren, T.A.: Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17(5–6), 490–519 (1996). https://doi.org/10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P
Tosco, P., Stiefl, N., Landrum, G.: Bringing the MMFF force field to the RDKit: implementation and validation. J. Cheminformatics 6(1), 1–4 (2014). https://doi.org/10.1186/s13321-014-0037-3
Wang, S., Witek, J., Landrum, G.A., Riniker, S.: Improving conformer generation for small rings and macrocycles based on distance geometry and experimental torsional-angle preferences. J. Chem. Inf. Model. 60(4), 2044–2058 (2020). https://doi.org/10.1021/acs.jcim.0c00025
Gasteiger, J., Marsili, M.: A new model for calculating atomic charges in molecules. Tetrahedron Lett. 19(34), 3181–3184 (1978). https://doi.org/10.1016/S0040-4039(01)94977-9
Rappe, A.K., Casewit, C.J., Colwell, K.S., Goddard, W.A., Skiff, W.M.: UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 114(25), 10024–10035 (1992). https://doi.org/10.1021/ja00051a040
O’Boyle, N.M., Banck, M., James, C.A., Morley, C., Vandermeersch, T., Hutchison, G.R.: Open babel: an open chemical toolbox. J. Cheminformatics 3(1), 33 (2011). https://doi.org/10.1186/1758-2946-3-33
MGLTools Homepage. http://mgltools.scripps.edu/. Accessed 10 Dec 2021
Alhossary, A., Handoko, S.D., Mu, Y., Kwoh, C.-K.: Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics 31(13), 2214–2216 (2015). https://doi.org/10.1093/bioinformatics/btv082
Trott, O., Olson, A.J.: AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31(2), 455–461 (2010). https://doi.org/10.1002/jcc.21334
Hochreiter, S., Schmidhuber, J.: Long short-term memory. Neural Comput. 9(8), 1735–1780 (1997). https://doi.org/10.1162/neco.1997.9.8.1735
TensorFlow Homepage. https://www.tensorflow.org/. Accessed 10 Dec 2021
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: Proceedings of the 3rd International Conference on Learning Representations (ICLR), San Diego (2015)
Acknowledgments
This study was financed by grants of the Belarusian Republican Foundation for Fundamental Research (projects F21COVID-002 and F21ARMG-001) with the support of the Alliance of International Organizations (ANSO-CR-PP-2021-04). The authors are also grateful to the PRIP2021 Conference team for the selection of this study to be supported for publication.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Shuldau, M.A., Yushkevich, A.M., Bosko, I.P., Tuzikov, A.V., Andrianov, A.M. (2022). Generative Autoencoders for Designing Novel Small-Molecule Compounds as Potential SARS-CoV-2 Main Protease Inhibitors. In: Tuzikov, A.V., Belotserkovsky, A.M., Lukashevich, M.M. (eds) Pattern Recognition and Information Processing. PRIP 2021. Communications in Computer and Information Science, vol 1562. Springer, Cham. https://doi.org/10.1007/978-3-030-98883-8_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-98883-8_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-98882-1
Online ISBN: 978-3-030-98883-8
eBook Packages: Computer ScienceComputer Science (R0)