Abstract
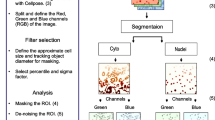
Fluorescence microscopy has enabled imaging of spatial proteome (morphological pattern of subcellular protein localization). Automated (and even manual) high-resolution fluorescence image acquisition generates a large amount of complex image data, and manual analysis of such data in order to distill biologically meaningful information is challenging. Automated image analysis is an inevitable approach to perform quantification of phenotypic changes, evade subjective bias, and provide accurate and reproducible results. Automation requires the utilization of image processing, image analysis, and data analysis tools. In Chap. 9 is presented our customized system for automated analysis of fluorescently stained blood cells. Our aim was to develop an automatic image analysis system that would enable us to minimize the amount of required manual operations not only throughout the inspection of segmentation results but also during the initial tuning of various parameters of the segmentation algorithm. Setting free from the tuning of initial parameters allows for faster switching to data analysis of the new experiment when imaging settings or other conditions were changed. The development of the fluorescence image analysis algorithms included a good practice from the development of 2DE image analysis system (see Chap. 6). The developed tools were applied for automated analysis of confocal microscopy images to evaluate changes of histone modifications in cell populations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Andrade AR, Vogado LHS, de M S Veras R, Silva RRV, Araujo FHD, Medeiros FNS (2019) Recent computational methods for white blood cell nuclei segmentation: a comparative study. Comput Methods Programs Biomed 173:1–14
Antony PPMA, Trefois C, Stojanovic A, Baumuratov AS, Kozak K (2013) Light microscopy applications in systems biology: opportunities and challenges. Cell Commun Signaling 11(1):24
Bajcsy P, Chalfoun J, Simon M (2018) Web microanalysis of big image data. Springer, New York
Bankman I (2008) Handbook of medical image processing and analysis, 2nd edn. Elsevier, Amsterdam
Bashar MK, Komatsu K, Fujimori T, Kobayashi TJ (2012) Automatic extraction of nuclei centroids of mouse embryonic cells from fluorescence microscopy images. Plos One 7(5):e35550
Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, Zhang C, Koethe U, Hamprecht FA, Kreshuk A (2019) Ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16(12): 1226–1232. https://doi.org/10.1038/s41592-019-0582-9
Buades A, Coll B, Morel JM (2005) A review of image denoising algorithms, with a new one. Multiscale Model. Simul. 4(2):490–530
Cao H, Liu H, Song E (2018) A novel algorithm for segmentation of leukocytes in peripheral blood. Biomed Signal Process Control 45:10–21
Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006) Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7(10):R100
Chiarini-Garcia H, Melo RCN (2011) Light Microscopy. In: Methods and Protocols. Springer, New York
Choudhry P (2016) High-throughput method for automated colony and cell counting by digital image analysis based on edge detection. PloS one 11(2):e0148469. https://doi.org/10.1371/journal.pone.0148469
Collins A, Huett A (2018) A multi-phenotypic imaging screen to identify bacterial effectors by exogenous expression in a HeLa cell line. Sci Data 5(1):1–12. https://doi.org/10.1038/sdata.2018.81
Courtney J, Woods E, Scholz D, Hall WW, Gautier VW (2015) MATtrack: a matlab-based quantitative image analysis platform for investigating real-time photo-converted fluorescent signals in live cells. PloS one 10(10):e0140209
De Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, et al. (2012) Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9(7):690–696
Domínguez C, Heras J, Pascual V (2017) IJ-OpenCV: Combining ImageJ and OpenCV for processing images in biomedicine. Comput Biol Med 84:189–194
Dukes KA, Sullivan LM (2007) A review of basic biostatistics. Evaluating Techniques in Biochemical Research Cell Press, Cambridge, MA, pp 50–56
Elen A, Turan MK (2018) A new approach for fully automated segmentation of peripheral blood smears. Int J Adv Appl Sci 5(1):81–93
Eliceiri KW, Berthold MR, Goldberg IG, Ibáñez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, et al. (2012) Biological imaging software tools. Nat Methods 9(7):697
Fouad S, Landini G, Randell D, Galton A (2016) Morphological separation of clustered nuclei in histological images. In: International Conference on Image Analysis and Recognition. Springer, New York, pp 599–607
Furtado A, Henry R (2002) Measurement of green fluorescent protein concentration in single cells by image analysis. Anal Biochem 310(1):84–92
Gomes-Alves AG, Maia AF, Cruz T, Castro H, Tomas AM (2018) Development of an automated image analysis protocol for quantification of intracellular forms of Leishmania spp. PloS one 13(8):e0201747. https://doi.org/10.1371/journal.pone.0201747
Gonzalez RC, Woods RE (2018) Digital Image Processing, 4th edn. Prentice Hall, Englewood Cliffs
Gudla PR, Nandy K, Collins J, Meaburn KJ, Misteli T, Lockett SJ (2008) A high-throughput system for segmenting nuclei using multiscale techniques. Cytometry Part A J Int Soc Anal Cytol 73(5):451–466
Haidekker M (2010) Advanced biomedical image analysis. Wiley, New York
Hawkes PW, Spence JCH (2019) Springer handbook of microscopy. Springer, New York
Hegde RB, Prasad K, Hebbar H, Sandhya I (2018) Peripheral blood smear analysis using image processing approach for diagnostic purposes: A review. Biocybern Biomed Eng 38(3):467–480
Hegde RB, Prasad K, Hebbar H, Singh BMK (2019) Development of a robust algorithm for detection of nuclei of white blood cells in peripheral blood smear images. Multimedia Tools Appl 78(13):17879–17898
Held M, Schmitz MHA, Fischer B, Walter T, Neumann B, Olma MH, Peter M, Ellenberg J, Gerlich DW (2010) Cellcognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat Methods 7(9):747–754
Hodneland E, Kögel T, Frei DM, Gerdes HH, Lundervold A (2013) Cellsegm-a MATLAB toolbox for high-throughput 3D cell segmentation. Source Code Biol Med 8(1):16
Hussain A, Ghosh S, Kalkhoran SB, Hausenloy DJ, Hanssen E, Rajagopal V (2018) An automated workflow for segmenting single adult cardiac cells from large-volume serial block-face scanning electron microscopy data. J Struct Biol 202(3):275–285
Jerome WG, Price RL (2018) Basic confocal microscopy. Springer, New York
Johnson HJ, McCormick MM, Ibanez L (2015a) The ITK software guide book 1: introduction and development guidelines, vol 1. Kitware, Inc., New York
Johnson HJ, McCormick MM, Ibanez L (2015b) The ITK software guide book 2: design and functionality, vol 2. Kitware, Inc., New York
Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. (2009) Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci 106(6):1826–1831
Kaehler A, Bradski G (2016) Learning OpenCV 3: computer vision in C++ with the OpenCV library. O’Reilly Media, Inc., New York
Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE (2011) Improved structure, function and compatibility for cellprofiler: modular high-throughput image analysis software. Bioinformatics 27(8):1179–1180
Kankaanpää P, Paavolainen L, Tiitta S, Karjalainen M, Päivärinne J, Nieminen J, Marjomäki V, Heino J, White DJ (2012) Bioimagexd: an open, general-purpose and high-throughput image-processing platform. Nat Methods 9(7):683–689
Kapur JN, Sahoo PK, Wong AKC (1985) A new method for gray-level picture thresholding using the entropy of the histogram. Comput Vision Graphics Image Proc 29(3):273–285
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vision 1(4):321–331
Krzywinski M, Altman N (2013a) Points of significance: importance of being uncertain. Nat Methods 10(9):809–810
Krzywinski M, Altman N (2013b) Points of significance: significance, p values and t-tests. Nat Methods 10(11):1041–1042
Krzywinski M, Altman N (2014a) Points of significance: comparing samples–part i. Nat Methods 11(4):215–216
Krzywinski M, Altman N (2014b) Points of significance: visualizing samples with box plots. Nat Methods 11(2):119–120
Kubitscheck U (2017) Fluorescence microscopy: from principles to biological applications. Wiley, New York
Lamprecht MR, Sabatini DM, Carpenter AE (2007) Cellprofiler: free, versatile software for automated biological image analysis. Biotechniques 42(1):71–75
Lawlor D (2019) Introduction to light microscopy: tips and tricks for beginners. Springer, Berlin
Legland D, Arganda-Carreras I, Andrey P (2016) Morpholibj: integrated library and plugins for mathematical morphology with imagej. Bioinformatics 32(22):3532–3534
Li G, Liu T, Nie J, Guo L, Malicki J, Mara A, Holley SA, Xia W, Wong STC (2007) Detection of blob objects in microscopic zebrafish images based on gradient vector diffusion. Cytometry Part A J Int Soc Anal Cytol 71(10):835–845
Li F, Yin Z, Jin G, Zhao H, Wong STC (2013) Bioimage informatics for systems pharmacology. PLoS Comput Biol 9(4):e1003043
Ljosa V, Carpenter AE (2009) Introduction to the quantitative analysis of two-dimensional fluorescence microscopy images for cell-based screening. PLoS Comput Biol 5(12):e1000603
Lundberg E, Borner GH (2019) Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 20(5):285–302
Markaki Y, Harz H (2017) Light microscopy. In: Methods and protocols. Springer, New York
McCullough DP, Gudla PR, Harris BS, Collins JA, Meaburn KJ, Nakaya MA, Yamaguchi TP, Misteli T, Lockett SJ (2008) Segmentation of whole cells and cell nuclei from 3-d optical microscope images using dynamic programming. IEEE Trans. Med. Imaging 27(5):723–734
McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018) Cellprofiler 3.0: Next-generation image processing for biology. PLoS Biol 16(7):e2005970
Meijering E, Carpenter AE, Peng H, Hamprecht FA, Olivo-Marin JC (2016) Imagining the future of bioimage analysis. Nat Biotechnol 34(12):1250
Miura K, Sladoje N (2020) Bioimage data analysis workflows. Springer, Berlin
Model MA, Burkhardt JK (2001) A standard for calibration and shading correction of a fluorescence microscope. Cytometry J Int Soc Anal Cytol 44(4):309–316
Nandy K, Gudla PR, Meaburn KJ, Misteli T, Lockett SJ (2009) Automatic nuclei segmentation and spatial fish analysis for cancer detection. In: Proceedings of the 2009 annual international conference of the IEEE engineering in medicine and biology society. IEEE, New York, pp 6718–6721
Nandy K, Gudla PR, Amundsen R, Meaburn KJ, Misteli T, Lockett SJ (2012) Automatic segmentation and supervised learning-based selection of nuclei in cancer tissue images. Cytometry Part A 81(9):743–754
Navakauskiene R, Borutinskaite VV, Treigyte G, Savickiene J, Matuzevicius D, Navakauskas D, Magnusson KE (2014) Epigenetic changes during hematopoietic cell granulocytic differentiation—comparative analysis of primary CD34+cells, KG1 myeloid cells and mature neutrophils. BMC Cell Biol 15:4. https://doi.org/10.1186/1471-2121-15-4
North AJ (2006) Seeing is believing? a beginners’ guide to practical pitfalls in image acquisition. J Cell Biol 172(1):9–18
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Ouyang W, Winsnes CF, Hjelmare M, Cesnik AJ, Åkesson L, Xu H, Sullivan DP, Dai S, Lan J, Jinmo P, et al. (2019) Analysis of the human protein atlas image classification competition. Nat Methods 16(12):1254–1261
Paulik R, Micsik T, Kiszler G, Kaszal P, Szekely J, Paulik N, Varhalmi E, Premusz V, Krenacs T, Molnar B (2017) An optimized image analysis algorithm for detecting nuclear signals in digital whole slides for histopathology. Cytometry Part A 91A(6, SI):595–608. https://doi.org/10.1002/cyto.a.23124
Pawley J (2006) Handbook of biological confocal microscopy, vol 236. Springer, New York
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 12(7):629–639
Phoulady HA, Goldgof D, Hall LO, Mouton PR (2017) A framework for nucleus and overlapping cytoplasm segmentation in cervical cytology extended depth of field and volume images. Comput Med Imaging Graphics 59:38–49
Pietzsch T, Preibisch S, Tomančák P, Saalfeld S (2012) ImgLib2–generic image processing in Java. Bioinformatics 28(22):3009–3011
Prinyakupt J, Pluempitiwiriyawej C (2015) Segmentation of white blood cells and comparison of cell morphology by linear and naïve bayes classifiers. Biomed Eng Online 14(1):63
Qiao G, Zong G, Sun M, Wang J (2012) Automatic neutrophil nucleus lobe counting based on graph representation of region skeleton. Cytometry Part A 81(9):734–742
Ranefall P, Wahlby C (2016) Global gray-level thresholding based on object size. Cytometry Part A 89A(4):385–390. https://doi.org/10.1002/cyto.a.22806
Rebollo E, Bosch M (2019) Computer optimized microscopy: methods and protocols. Springer, New York
Rittscher J, Machiraju R, Wong STC (2008) Microscopic image analysis for life science applications. Artech House, New York
Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) Imagej2: imagej for the next generation of scientific image data. BMC Bioinf 18(1):529
Russ JC, Neal FB (2017) The image processing handbook, 7th edn. CRC Press, West Palm Beach
Sajjad M, Khan S, Jan Z, Muhammad K, Moon H, Kwak JT, Rho S, Baik SW, Mehmood I (2016) Leukocytes classification and segmentation in microscopic blood smear: a resource-aware healthcare service in smart cities. IEEE Access 5:3475–3489
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to imagej: 25 years of image analysis. Nat Methods 9(7):671–675
Schnipper N, Stassen HH, Kallinich T, Sperling K, Hoffmann K (2017) Image analysis of neutrophil nuclear morphology: learning about phenotypic range and its reliable analysis from patients with pelger-huët-anomaly and treated with colchicine. Cytometry Part B Clin Cytometry 92(6):541–549
Schroeder W, Martin K, Lorensen B (2006) The visualization toolkit, 4th edn. Kitware, New York
Schüffler PJ, Fuchs TJ, Ong CS, Wild PJ, Rupp NJ, Buhmann JM (2013) TMARKER: a free software toolkit for histopathological cell counting and staining estimation. J Pathol Inf 4(2):2
Schwarzfischer M, Marr C, Krumsiek J, Hoppe P, Schroeder T, Theis FJ (2011) Efficient fluorescence image normalization for time lapse movies. In: Proc. Microscopic Image Analysis with Applications in Biology. MIAAB
Seruca R, Suri JS, Sanches JM (2017) Fluorescence imaging and biological quantification. CRC Press, New York
Sethian JA (1999) Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science, vol 3. Cambridge University, Cambridge
Sluder G, Wolf DE (2013) Digital microscopy. Academic Press, New York
Soille P (2013) Morphological image analysis: principles and applications. Springer, New York
Sommer C, Straehle CN, Köthe U, Hamprecht FA (2011) Ilastik: interactive learning and segmentation toolkit. In: Proceedings of the 8th IEEE international symposium on biomedical imaging (ISBI 2011), pp 230–233. https://doi.org/10.1109/ISBI.2011.5872394, 1
Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017) TrackMate: an open and extensible platform for single-particle tracking. Methods 115:80–90
Tomasi C, Manduchi R (1998) Bilateral filtering for gray and color images. In: Sixth International Conference on Computer Vision (IEEE Cat. No. 98CH36271). IEEE, New York, pp 839–846
Usaj MM, Styles EB, Verster AJ, Friesen H, Boone C, Andrews BJ (2016) High-content screening for quantitative cell biology. Trends Cell Biol. 26(8):598–611
Van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T (2014) scikit-image: image processing in Python. Peer J 2:e453
Vincent L (1993) Morphological grayscale reconstruction in image analysis: applications and efficient algorithms. IEEE Trans Image Process 2(2):176–201
Vincent L, Soille P (1991) Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. (6):583–598
Wallace W, Schaefer LH, Swedlow JR (2001) A workingperson’s guide to deconvolution in light microscopy. Biotechniques 31(5):1076–1097
Wang Y, Cao Y (2019) Leukocyte nucleus segmentation method based on enhancing the saliency of saturation component. J. Algorithms Comput. Technol. 13:1748302619845783
Waters JC (2009) Accuracy and precision in quantitative fluorescence microscopy
Waters J, Wittmann T (2014) Quantitative imaging in cell biology. Academic Press, New York
Xue JH, Zhang YJ (2012) Ridler and calvard’s, kittler and illingworth’s and otsu’s methods for image thresholding. Pattern Recognit Lett 33(6):793–797
Yang SJ, Berndl M, Ando DM, Barch M, Narayanaswamy A, Christiansen E, Hoyer S, Roat C, Hung J, Rueden CT, Shankar A, Finkbeiner S, Nelson P (2018) Assessing microscope image focus quality with deep learning. BMC Bioinf 19(1):1–9. https://doi.org/10.1186/s12859-018-2087-4
Author information
Authors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Navakauskienė, R., Navakauskas, D., Borutinskaitė, V., Matuzevičius, D. (2021). Computational Methods for Protein Localization Analysis. In: Epigenetics and Proteomics of Leukemia. Springer, Cham. https://doi.org/10.1007/978-3-030-68708-3_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-68708-3_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-68707-6
Online ISBN: 978-3-030-68708-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)