Abstract
Conserving the genetic diversity of potato is critical for the long-term future of potato improvement programs. Further, it is the social and ethical responsibility of the present generation to ensure future generations have the same opportunities to use, exploit, and benefit from the genetic diversity that exists today. Genebanks and the ex situ conservation of potato genetic resources are the only way to ensure this happens; in situ conservation plays a complementary role, but it can never ensure that the vast diversity that exists on earth today is still there for use in the future. Material in ex situ genebanks not only serve as a reservoir of ready-to-use genetic material when needed but also provide invaluable tools for research now and in the future of cultivated potato and its wild relatives.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
1 Ex Situ Conservation of Potato
The conservation of crop diversity, such as potato diversity , outside of its natural habitat, or ex situ, is held in botanical gardens and genebanks throughout the world. While botanical gardens conserve diversity and display this diversity for the public to enjoy and learn from, as well for scientific research, the overall mandate of botanical gardens is not generally to promote the use of and share the plant diversity they hold. In contrast, the mandate of genebanks is to provide access to the genetic resources they hold and to promote the use of these genetic resources for training, breeding, and research. Unfortunately, many national genebanks holding potato germplasm do not have the capacity, resources, nor a national mandate to widely distribute their holdings, and thus, access to potato germplasm is mostly left to the genebanks in developing countries, United States Department of Agriculture (USDA), and the potato genebank of the CGIAR, which holds potato germplasm at the International Potato Center, or CIP after its Spanish acronym located in Lima, Peru.
Potato genetic resources held ex situ are divided into potato wild relatives (maintained as seed populations) and cultivated varieties which are maintained either in field plantings (where tubers are planted and harvested annually), seed, in vitro/clonal material, or cryopreserved material. The same genetic material could be held in two or more different forms in a single genebank (in vitro and cryopreserved) or between two genebanks (seed and in vitro). What is critical for a genebank, is that the material it holds is available in a form that can be readily used by all who need it, including breeders, large and small-holder farmers, industry, researchers from developing or developed countries, and for teaching and educational purposes. Thus, the wealth of potato diversity (Figs. 4.1, 4.2, 4.4, and 4.5) held and made available by genebanks is an asset for humanity, as it safeguards known genetic traits which are useful today, along with a myriad of known and unknown traits ready to be deployed to meet the needs for the threats and challenges potato farmers face in the future.
The more uniform the crop in general, the easier it is to manage as an ex situ collection. Unfortunately, cultivated potato is an incredibly diverse crop, and while virtually all the potato planted and used commercially worldwide are tetraploid potato varieties, diploid, triploid, tetraploid, and even pentaploid landrace potatoes are still grown throughout the South American Andes. This diversity has been collected, maintained and is available for use in breeding and research programs worldwide from numerous genebanks. The diversity of cultivated potato is reflected not only in its vast ploidy range but also at the species level where over 30 years ago cultivated potato was divided into seven species and nine taxa [Solanum tuberosum (C. Linneo) subsp. andigenum (Juz. & Bukasov), S. tuberosum (C. Linneo) subsp. tuberosum, S. xchaucha (Juz & Bukasov), S. stenotomum subsp. goniocalyx (Juz. & Bukasov), S. stenotomum subsp. stenotomum (Juz. & Bukasov), S. xjuzepczukii (Bukasov), S. phureja (Juz. & Bukasov), S. xajanhuiri (Juz. & Bukasov), and S. xcurtilobum (Juz. & Bukasov); (Hawkes 1990)] (Figs. 4.1 and 4.2). More recently the taxonomy of potato has been revised and cultivated potato has been regrouped into four species (Solanum tuberosum, S. xjuzepczukii, S. xajanhuiri, and S. xcurtilobum; Spooner et al. 2014). A similar taxonomic revision has occurred in the taxa of potato wild relatives with the previous taxonomic classification of Hawkes (1990) recognizing 228 wild species in 21 series. However, the recent taxonomic classification by Spooner et al. (2014) reduced the wild relatives of potato (Solanum section Petota) to 107 species.
The change in taxonomy has created a split in the ex situ potato world with some genebanks (such as the International Potato Center (CIP)] still organizing their collection based on the taxonomy of Hawkes (1990) while other major potato genebanks [such as the United States Department of Agriculture-Agricultural Research Service (USDA-ARS)] adopting the taxonomy of Spooner et al. (2014). Potato curators in all genebanks acknowledge the revision by Spooner et al. (2014) as needed and an advance in the field, yet the groupings by Hawkes (1990) make sense for managing an ex situ collection. Spooner et al. (2014) foresaw such challenges in their revision of the potato taxonomy and therefore stated that due to the “complicating biological factors in section Petota”, they consider their “taxonomy to be subject to critique and modification” (Spooner et al. 2014, p 325). Two final comments are that there is consensus that ploidy in potato is not a good factor in delimitating species boundaries and that the use of different taxonomic names for the same material in different major potato genebanks is a detriment to use of the materials for the potato research community. That said, harmonizing the taxonomy used in global potato genebanks will need to be a focus in the next decade along with an extensive genetic comparison of potato collections to identify unique and redundant material between genebanks.
Taxonomic differences aside, ex situ collections and genebanks continue to play an ever increasingly dynamic role in providing the tools needed for food security in the future. The Second Report on The State of the World’s Plant Genetic Resources for Food and Agriculture (FAO 2010) estimates that there are over 1750 genebanks worldwide holding approximately 7.4 million accessions; however, the report estimates that only 25–30% of these accessions are genetically unique. The report lists 98,285 potato accessions conserved in 174 genebanks around the world, and using the estimate above of the percentage of unique accessions, we can assume that there are at most an estimated 24,500–29,500 unique potato accessions are conserved worldwide in genebanks. In the case of potato, the accuracy of such an estimate of unique accessions is difficult to access until a thorough rationalization-comparison amongst the potato ex situ collections is carried out. According to this same report, 6 genebanks hold 41% of the global potato accessions: The French National Institute for Agricultural Research (INRA) in France (11%), Vavilov Institute in Russia (9%), The International Potato Center (CIP) in Peru (8%), The Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Germany (5%), USDA-ARS in USA (5%), and The National Institute of Agrobiological Sciences (NIAS) in Japan (3%) and 20 global genebanks hold over 1000 potato accessions each. The listed global potato holdings collectively consist of 15% wild relatives of potato, 20% cultivated potato, 16% research and breeding materials, 14% advanced breeding lines, and 35% uncategorized accessions in these genebanks.
The last Global Strategy for Ex Situ Conservation of Potato (2006) analyzed 23 global potato collections which collectively maintain “nearly 59,000” potato accessions. This summary states that the genebanks in Latin America contain principally native cultivars while those in Europe and North America contain modern cultivars, breeding materials, and wild relatives. Such generalizations oversimplify the collections and indeed, although there is specialization among the potato genebanks, most major collections have a good representation of both wild and cultivated accessions.
Information about the particular attributes for each individual accession is critical for users to select which one or two of the thousands of accessions contain the trait(s) the user needs. Unfortunately, the information on accessions in global potato genebanks is generally incomplete and lacking the trait information users want and need most. Further, information on qualitative, complex traits (i.e., drought, frost tolerance, or yield) can be very specific to the physiological age at which the plant is subjected to the stress, the location, soil type, and degree of stress and, thus, is not always easy to list in a genebank database. Further, these complex traits require multiyear, multilocation evaluations in order to understand the complexities of the traits and genes associated with the trait. The USDA-ARS collection contains a large list of descriptors and traits recorded with 137 morphological, biochemical, nutritional, or physiological traits listed for potato (https://npgsweb.ars-grin.gov/gringlobal/cropdetail.aspx?type=descriptor&id=73), while CIP lists 72 descriptors or traits for their potato holdings (http://genebank.cipotato.org/gringlobal/search.aspx) and the European Union Genetic Resources of Potato lists 58 descriptors or traits (http://ecpgr.cgn.wur.nl/eupotato/). All these data are accessible in English on public internet-accessible databases, as is information from other global potato genebanks; although far too much of the information of the potato ex situ collections is not available publicly and outside of a few genebanks, much of the available listings from genebanks contain little information about the accessions. Fortunately, efforts such as Genesys , the global Gateway to Genetic Resources (https://www.genesys-pgr.org/welcome), provides descriptor and trait information from multiple genebanks in a single database, which facilitates use by facilitating the location and finding information on potato accessions worldwide.
2 Collection of New Potato Germplasm
There are estimations that 20% of plant species are in danger of extinction (Jansky et al. 2013) and such losses will be accelerated by climate change and habitat destruction. Further, for some plants, a large amount of their diversity may already only be able available from genebanks (Jarvis et al. 2011), which might be the future for potato where the wild relative species exist only in the fragile ecosystems of the Andes (Hijmans and Spooner 2001), where a changing climate is already having a substantial impact. Lingering questions for ex situ and in situ management of potato revolve around how to define diversity there is/was in cultivated and the large secondary genepools (wild potato relatives) of potato. For conservationists, the big question is how much of the diversity in potato is securely conserved, what diversity has been lost or is in imminent danger of being lost, and what is the economic value, as well as potential future value, of the potato diversity that is not securely conserved.
A benchmark study (Castañeda-Álvarez et al. 2015) evaluated the status of ex situ collections for 73 species (according to Spooner et al. 2014) of potato wild relatives and used environmental niche modelling (ENM) techniques to estimate potential geographic ranges of each species. Their data assigned a high priority status for collecting 32 species of wild potato, 43.8% of the species studied, due to severe gaps in the ex situ collections. Further, 20 more species were assigned medium priority for collection and only three species were determined to have good diversity representation in the ex situ collections. As part of the study, they also looked at in situ sites which were potentially threatened including that of S. rhomboideilanceolatum whose native habitat in Peru is increasingly threatened by road building and overgrazing. Potato wild relatives have made critical contributions to potato disease resistance, enhanced yield, and improved quality in past 50 years (Jansky et al. 2013). Therefore, it should be the obligation of present day potato researchers and breeders to do all we can to leave future potato scientists the same opportunities to explore, use, and reap benefits from the diversity of wild potato that we have today.
The geographic range of wild potato species is broadly spread out in 16 countries confined to the Americas (Mexico, United States, Costa Rica, Guatemala, Honduras, Panama, Argentina, Bolivia, Brazil, Chile, Colombia, Ecuador, Paraguay, Peru, Uruguay, and Venezuela (Fig. 4.3). For thousands of years, the ecological niches of the Andes have been the natural habitat and the center of the genetic diversity of wild potato species and innumerable native varieties. Long evolutionary processes have allowed the accumulation of genetic components that are valuable resources for the improvement of potato cultivation, such as the ability to survive adverse biotic conditions (diseases, pests) and abiotic (drought, frost, climate change), as well as, vast morphological differences (Figs. 4.4 and 4.5). However, this valuable genetic diversity is threatened by “genetic erosion.” The main threats are urbanization, alternative land use, and the destruction of natural areas. Examples include overgrazing, the construction of new routes and villages, and the destruction of forests.
A map of the distribution of wild potato accessions in the CIP genebank. Greens dot indicates origin of the accession based on passport data (https://cipotato.org/genebankcip/process/potato/potato-wild/)
The domestication of potato is thought to have occurred ~10,000 years ago in the Lake Titicaca Basin on the border between Peru and Bolivia, most likely from the S. brevicaule complex with S. candolleanum as a potential ancestor (Spooner et al. 2014). The use of these wild potato species for food is likely, yet the high glycoalkaloid content needed to be dealt with in some fashion to avoid toxicity. Rumold and Aldenderfer (2016) studied early potato starch grains from tools found at a village site in the Titicaca basin (1700–3500-year-old site) which resembled freeze-dried samples rather than starch from fresh tubers. They hypothesize that a process such as drying and grinding may have played a role in the detoxification of the wild potato tubers, as in other root crops. This theory is consistent with the natural freeze drying of tubers in the Andes today known as “chuño.” Recent supporting evidence for use of wild potatoes for human consumption comes from S. jamesii starch grains found among other 6000-year-old early human ruins in the Southwest US (Louderback and Pavlik 2017). It is interesting to note that S. jamesii was still used until recently as a food source for native communities in the area and that existing patterns of present day occurrence of this species suggests that early humans may have cultivated the tubers (Kinder et al. 2017). Domestication is believed to have given rise to increased tuber size through selection and cultivation which was associated with an increase in leaf carbon fixation and transport, reduction in glycoalkaloids, adaptation to long-day photoperiod, and reduced sexual fertility (Ryoko 2015).
With domestication of early edible potato tubers, it is logical this also was the start not only of selection and cultivation but also of movement of selected potato tubers from one site to another. As early as 5000 years ago, maize, originating in Mexico, was already in S. America (Grobman et al. 2012), hence there is no reason to think that the north to south movement of food materials was not also reciprocated with a south to north movement of food such as cultivated potato. The movement of domesticated varieties most certainly also gave rise to the movement of new genetic material through the zone of native wild populations followed by intercrossing leading to increased diversity in the domesticated varieties and further selection for adaptive traits. These same adaptive traits and the diversity associated with them contributed to the adaptations which were selected for by early human populations and these same traits still exist in the wild populations today and will continue to be of critical value to breeders as long as potato improvement programs exist.
Thus collecting, movement, and exchange of potato and potato wild relatives has occurred for millennia, yet was mainly confined to the Americans until the Spanish conquest when potato was collected and shipped to Europe where it underwent further selection. Wild relatives were likely also collected before the last century, but reliable documentation of the collection of wild potato for genebanking is more recent. Darwin, in his famous voyage of the Beagle, collected potato from the Chilóe Islands in Chile in the 1830s which he named S. maglia, now known as S. tuberosum (Ristaino and Pfister 2016). At this time, Darwin erroneously assumed potatoes originated from these islands due to the diversity and wide spread presence. In his journals, he referenced that the crew “stocked up on game and potatoes.” Fuentes (2014) references collection expeditions as yearly as the 1830–1833 by A. D’Orbigny from France in Bolivia. Perhaps the best documented collections are those done by the Vavilov Institute (Supreme Council of People’s Economy) in Russia starting in 1925 by S.M. Bukasov and S.V. Juzepzuk (Loskutov 1999), after whom the wild potato species S. bukasovii and S. xjuzepczukii were named, respectively. Although detailed notes from these expeditions were never published, it is known that many species of wild potato were collected in Peru, Bolivia, and Chile (Ochoa 2004), and that some of this material still is maintained at the Vavilov Institute (Prof. N. Dzyubenko, personal communication).
Collections of wild potato continued by numerous collectors from Europe, Asia, North America, and Latin American institutions in an ad hoc manner until the early 1970s (Correll 1962; Hawkes 1990; Ochoa 1962) when CIP organized a meeting of experts to form and develop an International Germplasm Bank at CIP in Lima, Peru. Three Planning Conferences were held dealing with topics ranging from exploration, collection, conservation, and evaluation of potato genetic resources ending with the recommendation of collection strategies and to make the collected material available to researchers for their use for the benefit of humanity (International Potato Center 1973–1974, 1976, 1979). In the last 50 years, CIP has organized or been involved in ~300 systematic exploration missions for potato. These included the collection of wild and native potato germplasm varieties under the direction of C.M. Ochoa and collaborating with some 245 national and international scientists including A. David, J.B. Bamberg., J.G. Hawkes, A.M. Van Harten, J.P. Hjerting, W. Hondelmann, R. Hoopes, K.A. Okada, A. Salas, D. Spooner, and J.J.C. Van Soest. These collecting missions organized from Peru, where the greatest genetic diversity of the Solanum section Petota (Solanaceae) wild and cultivated exists, included exploring and collecting in the 16 countries of the American continent. Numerous accessions of each of the wild potato species were collected in the unique ecological niches along the geographical distribution of each species. Of interest was the rediscovery of wild species previously described, but known only as herbarium specimens in addition to the discovery of about 20 new species (including, S. amayanum (Ochoa 1989a, b, c), S. bill-hookeri (Ochoa 1988a, b), S. bombycinum (Ochoa 1983a, b), S. chilliasense (Ochoa 1981a, b, c, d), S. incasicum (Ochoa 1981a, b, c, d), S. irosinum (Ochoa 1981a, b, c, d), S. longiusculus (Ochoa 1987), S. neovavilovii (Ochoa 1983a, b), S. orocense (Ochoa 1980a, b, c), S. ortegae (Ochoa 1998), S. peloquinianum (Ochoa 1980a, b, c), S. salasianum (Ochoa 1989a, b, c), S. sarasarae (Ochoa 1988a, b), S. simplicissimum (Ochoa 1989a, b, c), S. sucubunense (Ochoa 1980a, b, c), S. tapojense (Ochoa 1980a, b, c), S. taulisense (Ochoa 1981a, b, c, d)) which were characterized and described by Ochoa (Hawkes 1989, 1990; Ochoa 1962, 1990, 1999; Spooner et al. 1999, 2001). These accessions are all documented with full passport data (country and location of collection) and many are available for use globally through the CIP website (https://cipotato.org/genebankcip/process/potato/potato-wild/).
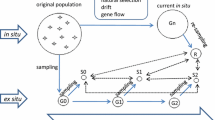
Most collection missions only have resources to focus collections from a single site and rarely can make multiple visits to the same site. Therefore, collectors are at the mercy of annual changes in rainfall and other abiotic and biotic factors that affect flowering or tuber germination. Exceptions are the collection trips by Bamberg and del Rio (2011) and Bamberg et al. (2003) where annual expeditions have been made to monitor in situ sites of S. jamesii and S. stoloniferum (formerly S. fendleri) since 1992 with over 200 visits. The expeditions were initiated to recollect from sites originally sampled in 1958 and 1978 to evaluate and characterize what changes could be measured over a 40+ year period and to maximize the ex situ collection of diversity found in this species (del Rio et al. 2001). Their research has yielded insight into the collection from wild potato, and perhaps, upset some long-held dogma. For example, as expected, they found more diversity using random amplified polymorphic DNAs (RAPDs) in seed collections, compared with the collection of tubers from individual plants in S. jamesii, but the opposite phenomena was observed for S. stoloniferum, where collection of tubers had greater diversity than seeds (Bamberg et al. 2009). In another report (del Rio et al. 1997), they found using RAPD markers, there were significant differences in allele frequencies between genebank-conserved material and recollected material from the same site in all seven comparisons of S. jamesii (diploid outcrosser), and 12 of 16 comparisons within S. stoloniferum (tetraploid inbreeder). While in situ biologists would conclude that this makes sense, since the argument is that evolutionary processes continue in situ, for the authors, this meant that recollecting from the same site was not only a good idea but also is desirable to capture new alleles (Bamberg et al. 2003). Other observations included a comparison of “easy” (road side collection sites) versus “remote” (sites one needed to hike to) where both collection methods yielded valuable alleles (Bamberg et al. 2010) and that patterns of geographic structure were not valuable predictors of genetic differences in these two species inferring that wide sampling of each population was needed to capture diversity.
3 International Fora for Plant Genetic Resources
The Convention of Biological Diversity (https://www.cbd.int/; CBD) is the most significant international treaty for the access to genetic resources globally; yet, it has had a negative impact on the collection of potato germplasm throughout the native range of wild potato. This is due to CBD reaffirming the sovereign rights of a nation to the genetic resources falling within the country boundaries, and hence, the country of origin has exclusive rights over terms for accessing genetic resources, including cultivated and wild relatives of potato, found within its boundaries. The treaty also reaffirmed the country of origin’s right to “fair and equitable sharing of benefits arising” from the use of their genetic resources. CBD further asserts that nations are responsible for the long-term protection and access to genetic resources over which they have jurisdiction. CBD also acknowledged that access to, and sharing of, genetic resources, and relevant technologies, are critical for meeting food and health needs of a growing world population. Thus, it is a country’s responsibility to not only protect but also make available for use and share in the benefits from such use of the genetic resources it holds. Unfortunately, most countries, and the international community generally, have been ill prepared for the responsibility of regulating access and for ensuring the sharing of benefits by third-party use of genetic resources, such as germplasm related to cultivated and wild potato relatives. Mechanisms to grant permission for the collection of plant genetic resources have taken time to implement, and in the meantime the collecting of plant genetic resources ceased. The challenge has been how to make genetic resources available for use on the one hand, while on the other, to ensure equitable sharing of benefits from the commercial utilization of the genetic resources, or derivatives thereof, by multiple third-party stakeholders’ years later. Further, definition of the terms under which genetic resources could be accessed and benefits shared have been defined in the Nagoya Protocol on Access and Benefit Sharing (https://www.cbd.int/abs/, Nagoya). All potato species, except for S. phureja, are covered by the International Treaty for Plant Genetic Resources for Food and Agriculture (https://www.fao.org/plant-treaty/; ITPGRFA) which provides a legal framework to distribute the genetic material for research, training, and breeding with the acceptance of a Standard Material Transfer Agreement (SMTA) by the recipient.
Presently, there is a lack of a defined, functional mechanism for granting permission for collecting germplasm from the wild in most Latin American nations. The collection of potato germplasm for ex situ conservation in the countries where the clear majority of cultivated potato landraces and wild relatives originated and exist, has been virtually nonexistent since 2000. The exception is the United States of America (USA), which is not a party to CBD, and hence germplasm from potato wild relatives have continued to be collected in the USA (Bamberg et al. 2003), although only two species [S. stoloniferum (previously S. fendleri) and S. jamesii] are confirmed to exist within the boundaries of the USA. In 2016, a significant change occurred when the Peruvian National Institute for Innovative Agriculture (Instituto Nacional de Innovacion Agraria, INIA) was granted a permit for the collection of potato wild relatives within Peru, and in 2017 three collection missions were carried out collecting more than 70 populations of 14+ species (putatively S. acaule, S. acroscopicum, S. aymaraesense, S. bukasovii, S. coelestispetalum, S. marinasense, S. pillahuatense S. raphanifolium, S. sandemani, S. sparsipilum, S. tacnaense, S. urubambae, S. velardei, S. yungasense) under the auspices of the ITPGRFA. It is hoped that this will provide a model for further collections of wild and cultivated potato within its native range.
The collection of potato in its native range should be facilitated by the inclusion of cultivated potato (except for S. phureja) and all wild potato species in the list of Annex 1 crops under the ITPGRFA. The ITPGRFA established a global Multilateral System (MLS) that clearly defines the terms under which access for the use of genetic resources and their derivatives would be granted for use in research, training, and breeding under a Standard Material Transfer Agreement (SMTA) (http://www.fao.org/plant-treaty/areas-of-work/the-multilateral-system/the-smta/en/), which clearly outlines benefit sharing options. Of nations where wild potato species are native, only Colombia and Mexico are not parties to ITPGRFA, and thus, it is hoped that further facilitated access to collecting, conservation, and use of cultivated and wild potato species for research, training, and breeding will commence. For Colombia and Mexico, collecting, access, and use of native potato germplasm would be subject to bilateral agreements under the Nagoya protocol framework. To our knowledge, no other collections of wild potato species for inclusion in the MLS of the ITPGRFA have been carried out since the 1990s.
4 Methods of Conservation of Potato Germplasm
A major challenge for ex situ genebanks is to maintain the diversity held, often thousands of different accessions, in a form where every accession is readily available and accessible for use at a moment’s notice. Collections of cultivated crops are often composed of various traditional varieties, along with improved cultivars, which often do not produce large quantities of seeds/tubers making them challenging to reproduce in field conditions. Additionally, in general, wild species are more challenging for ex situ genebanks to regenerate than cultivated due to the multitude of ecological niches they have become adapted to, often requiring unique environmental conditions (i.e., photoperiods) to thrive and reproduce, along with some genotypes/species, which genetically produce limited flowers, seeds, or tubers.
With the lifespan of most research grants being less than 5 years, delays in obtaining germplasm can mean missing critical planting windows which could result in a year’s delay or failure to meet deliverables for a project. Thus, availability of material from the genebanks is critical. Most crop genebanks maintain their germplasm collections as botanical seed which for the majority of plants, including potato, stores very well and can be made available with a few weeks’ notice if adequate seeds exist for that accession. Genebanks will only provide seed to requestors if they have adequate seed in cold storage with good viability to ensure that the material is safeguarded and there is enough seed to keep regenerating the accession. However, unlike most crops, cultivated potato is also often maintained as clones to preserve the unique allelic combinations in the selected material which poses additional challenges for maintenance and distribution.
Where desirable allelic combinations have not been fixed by selection, as in wild potato germplasm, the collections are maintained as botanical seed or true-potato-seed (TPS) . A representative number of individuals (20–50) are typically collected from a wild population and seed is regenerated and combined to form a unique genebank accession of heterogeneous seed theoretically representative of the alleles found in that population. The accession is then distributed as a population of 50–100 seed where each seed represents a distinct genotype from that population. Thus, what is distributed is a heterogeneous mix of genotypes (seed) with each genotype representing a portion of the genetic make-up of the accession or population. Different collections of the same species (individual accessions) may or may not contain the same trait(s) desired by a breeder or even have this trait at the same frequency in different seed populations distributed. For example, when assessing late blight (Phytophthora infestans) resistance in wild potato accessions, Perez et al. (2001) found very high uniform resistance in three accessions of S. orophilum (100%, 98%, and 96% resistant individuals, respectively from 48 seedlings from each accession); whereas, in three accessions of S. wittmachii, resistance was high in one accession but very low in another (75%, 46% and 0.02% resistant individuals, respectively from 48 seedlings from each accession).
Unlike the wild species, cultivated potato is generally maintained as clones; however, in some cases seed populations of cultivated material may be just as useful to users. One of the main advantages of maintaining cultivated potato as seed is the overall cost and staffing required to maintain a seed collection is greatly reduced compared to in vitro collections which require subculturing every year or two. Further, sterile laboratory conditions with specialized equipment are not needed to reproduce potato if maintained as seed rather than in vitro clones. Seed populations can be produced and if adequate seed is obtained, a curator would not need to regenerate the accession for several decades unless the vigor of the seed decreases or the seeds are exhausted. Additionally, seed populations would have more diversity and new allele combinations that could lead to new trait discovery.
In contrast to maintenance of cultivated material as seed, maintenance of cultivated potato as a clone, preserving the unique allelic makeup of the landrace or variety, has advantages for either direct planting or facilitating the transfer of unique allelic combinations to breeding or research programs. Maintenance of clonal potato collections can be in the field, greenhouse, or in vitro as tissue culture. If maintained in the field or greenhouse, the collection is harvested, tubers stored and annually replanted as maintaining tuber viability for longer than a year is unreliable. Field or greenhouse maintenance of the collection also carries a risk of losing accessions due to biotic (insects and diseases) or abiotic (hail or wind) challenges. Therefore, many clonal potato collections are maintained in tissue culture as in vitro plantlets, which virtually eliminates the challenges of external biotic and abiotic factors. In vitro maintenance also facilitates international distribution of the collection as tissue culture plants are maintained in a sterile environment, and once cleaned of diseases, they can be certified as disease-free material for import purposes internationally.
The cultivated potato germplasm collection at CIP originated in the early 1970s with material collected or donated from around the world. In the early 1980s, research was initiated to place the collection into in vitro culture. Few in vitro collections have been maintained for 30 years or that have been used as extensively as the CIP cultivated potato collection. CIP currently maintains the largest in vitro potato collection with 8354 potato in vitro accessions (as of October 2018), the majority of which (89.8%) are landraces (native potatoes or “papa nativas” in Spanish) originating mainly from the Andean region, with the remaining accessions being improved varieties and breeding lines.
Since the vast majority of potato germplasm in genebanks around the world was collected over 20 years ago, and some, such as the collection at the Vavilov Institute in Russia, almost 90 years ago, most potato ex situ collections would be virtually impossible to replicate or replace today if lost. For this reason, Standards 4.9 of the Genebank Standards for Plant Genetic Resources for Food and Agriculture (FAO 2014) clearly state that “A safety duplicate sample for every original accession should be stored in a geographically distant area, under the same or better conditions than those in the original genebank.” CIP, along with other genebanks in the CGIAR, whose collections are held in trust under the ITPGRFA, takes this a step further with a guideline for backing up the collection in two distinct locations, one nationally and a second backup internationally. In the case of TPS accessions, such as the wild relatives of potato, a sealed package of seed equivalent to one to three times what is needed for a regeneration are backed up in a remote island archipelago in Norway at the Svalbard Global Seed Vault (https://www.nordgen.org/sgsv/, SGSV). The SGSV was established 10 years ago and funded by the Norwegian government with operations coordinated by the Nordic Gene Resource Centre and the Global Crop Diversity Trust. Shipment and coordination of all seed deposits are done by the Nordic Gene Resource Centre under a MTA which clearly states that the objective of the vault is “…to provide a safety net for the international conservation system of plant genetic resources, and to contribute to securing the maximum amount of plant genetic diversity of importance to humanity for the long-term….” (https://www.nordgen.org/sgsv/index.php?page=welcome). Under this agreement, the material is kept by the SGSV as a black-box safety back-up which denotes that the depositor retains full ownership and rights to the material, with the back-up facility holding the material only as a service of storage for the depositor. As of the end of 2017, SGSV contained almost 900,000 samples of seed from different crops including seed from 136 species of wild potato, five wild potato natural hybrids and six species of cultivated potato (taxonomy according to Hawkes 1990). Under conditions at the SGSV (airtight containers, properly dried seed, stored at −18 °C), good quality TPS can survive and maintain viability for 50–100 years.
Unfortunately, there are no analogous safety backup facilities to the SGSV for clonal material. In the case of clonal collections maintained in the field, the expense is often too high to allow distinct duplicate field planting sites, and hence, these collections are all too often not backed up and diminish over time. In the case of in vitro collections, the collections can be safely backed-up by shipping duplicate copies of in vitro cultures to a distant location for safety back-up. The facility backing-up the in vitro material generally needs to have lighted controlled temperature chambers (5–10 °C) for sustained slow growth of the material. In the case of the back-up for the USDA National Potato Genebank in vitro collection, cultures were shipped to the National Laboratory for Genetic Resources Preservation in Fort Collins, CO, where they are maintained as mini-tubers at 5 °C requiring subculture and reinitiating of the mini-tubers once every 2–3 years. For other in vitro potato collections, such as the one at CIP, the entire collection is shipped periodically (semi-annually) to two sites, one in a geographically distinct location in the high Andean Plateau at a CIP field station in Huancayo, Peru and the other facility outside of Peru at the Brazilian Agricultural Research Corporation (EMBRAPA) labs in Brasilia, Brazil. At both sites, the material is kept in slow growth conditions at 7 °C where the cultures can survive approximately 2 years without subculturing. Again, as with the seed deposits at the SGSV, these materials are stored in a black-box arrangement.
As genebanks are the stewards of the genetic resources that they maintain, with this stewardship comes great responsibility to ensure that no known pathogens are introduced into other countries with the distribution of germplasm creating a new disease problem. Vegetatively propagated crops such as potato, sugarcane, cassava, sweetpotato, yam, banana, citrus, and strawberry can be challenging because they must be multiplied continuously when maintained in the field to keep them in optimal growth. Although field maintenance of any crop is subject to abiotic challenges, as well as, biotic challenges such as diseases and pests, and with each multiplication or movement from one field to another, there are ample opportunities for the introduction of viruses and viroids (Sastry and Zitter 2014). Diseases caused by plant viruses limit sustainable production of vegetatively propagated staple food crops and in developing countries, this problem is confounded as agronomic practices to limit disease incidence and spread is marginal if done at all. As well, the use of new clean planting material to lessen virus titers is a luxury which most small holder farmers cannot afford. Additionally, low virus titers in planting materials generally display low symptom severity which makes infected plants not easily recognized as disease containing, and subsequently problematic if used for vegetative propagation (Bosch et al. 2007) because it can further perpetuate the diseases. Often human actions and climate change can be linked to the spread and outbreak of disease in conjunction with the movement and exchange of diseased plants and the international food supply chain are drivers of new disease threats (Wilkinson et al. 2011).
Actual examples of the human spread of pathogens with potato germplasm, other than by word of mouth, are difficult to find, yet published reports do exist to verify the potential challenge. The recent introduction of East Africa cassava mosaic-like viruses from African to the South West Indian Ocean islands follows human and insect transmission of the disease (De Bruyn et al. 2012), while the spread of the disease to Oman is likely directly related to human movement of plant material in the 1960s (Khan et al. 2012). Spread of viruses across international borders can also happen by insect vectors as in the case of the expansion and subsequent pandemic of the cassava mosaic disease (CMD) in the Great Lakes region of East and Central Africa which followed tightly the outbreaks of whitefly populations (Legg et al. 2011).
Therefore, for international distribution of germplasm from genebanks, plant material is tested, treated to eliminate diseases and certified to be clean and free of pathogens of phytosanitary importance as defined by import permits prior to transporting across any international border. For virus-free production of TPS, regeneration is carried out under strict sanitary conditions in a controlled screen house with limited access and maintained free of insects and thus vectors for viruses. Arracacha virus B–oca (arracacha and oca are two Andean Root and Tubers crops cultivated and eaten throughout Peru) strain (AVB-O), potato virus T (PVT), potato spindle tuber viroid (PSTVd), and alfalfa mosaic virus (AMV) have been shown to transmit virus particles to seed when pollen from an infected plant is used for pollinations (Jones 1982; Valkonen et al. 1992; FAO/IPGRI Technical Guidelines for the Safe Movement of Germplasm n.d.)]. Therefore, at CIP, for individual TPS production mother plants are tested pre-flowering for these seed transmitted viruses (AVB-O, PVT, PSTVd, and AMV) as well as for Andean potato latent virus (APLV), potato yellowing virus (PYV), and tobacco mosaic virus (TMV). If an individual mother plant tests positive for any of the viruses listed above, it is discarded and not used as a pollen donor or as a maternal parent for TPS production.
Pathogen elimination from clonal material is much more laborious, costly, and time-consuming. This is one rationale for in vitro culture as once the plantlets are clean, the in vitro culture methodology will maintain the investment of the phytosanitary cleaning. If endogenous bacteria are present in the cultures, these are usually eliminated with antibiotics prior to the elimination of viruses. A typical and successful virus elimination scheme starts with placing the in vitro cultures into thermotherapy (32–34 °C for 1 month), followed by isolation of 0.1–0.3 mm meristems and then plantlet regeneration. The regenerated plants are tested for the presence of viruses by serological, molecular, host range testing, and grafting onto indicator plants for final assurance of virus-free status. The entire process takes 12–24 months for one round and longer if not successful on the first run. At CIP, this method is highly efficient with 90% of all potato material going through virus elimination becoming virus-free after one run. The viruses of import/export importance in clonal material include potato virus T (PVT), potato spindle tuber viroid (PSTVd), potato virus X (PVX), potato virus Y (PVY), potato leaf roll virus (PLRV), potato virus S (PVS), Andean potato mottle virus (APMV), Andean potato latent virus (APLV), potato yellowing virus (PYV), and Arracacha virus B (AVB-O).
Advances in next-generation sequencing (NGS) hold great promise for reducing the labor, time, and cost of disease screening in germplasm collections. One such technology is the detection of viruses using small RNA sequencing and assembly (sRSA) technology. This technology is being tested on potato and it is hoped to be able to reduce the time necessary for phytosanitary cleaning of clonal potato germplasm from 12–24 months to less than 6 months (Kreuze 2014). Importantly, this technology could be invaluable in the future as new viruses or diseases of import importance develop so that entire germplasm collections can be rescreened quickly and efficiently ensuring the germplasm is readily available when needed.
The maintenance of in vitro potato clonal material has its advantages, including being readily available and the maintenance of disease-free status, but it is also undisputedly one of the most expensive long-term conservation methods available. This is due to the need for sterile plant tissue culture facilities, climate-controlled growth rooms, and highly trained personnel. Maintenance of clonal material as seed is one alternative, yet to maintain the allelic combinations present in an accession, maintenance of the clone is the only option. Cryopreservation , freezing tissue in liquid nitrogen at −196 °C, has been used for the long-term preservation of clonal plant genetic resources. Although research in the cryopreservation of plants has been ongoing for over 50 years (Reed 2008) and high confidence exists with the technology in animal and human systems (Di Santo et al. 2012), the application of long-term preservation of plant material, despite many successes, still has its skeptics. Limited long-term studies of plant material have shown largely positive and optimistic results (Volk et al. 2008), which support the theoretical long-term cryopreservation of plant meristems for centuries. Potato was one of the early crop plants used in cryopreservation research (Bajaj 1977; Grout and Henshaw 1978; Towill 1981a, b) and there are reports of over 20 potato species having been tested for cryopreservation using several different methods (Kaczmarczyk et al. 2011; Vollmer et al. 2017). To date the most successful methods for potato cryopreservation of genebank accessions included the IPK genebank using dimethyl sulfoxide (DMSO) droplet vitrification (Kaczmarczyk et al. 2011) with over 1000 cryopreserved accessions and the CIP genebank using a Plant Vitrification Solution (PVS2) droplet vitrification (Vollmer et al. 2017) method with over 2500 accessions cryopreserved as of the end of 2017.
Although the number of accessions or samples distributed from potato ex situ collections tells us little about how the germplasm was used or if it was useful at all, it does provide a measure of interest in the collections. In marketing, if a product is not available and there is little information on the attributes and benefits of the product, few will buy or order it. The same is true with germplasm collections, if the germplasm is not phytosanitary cleaned, in a form which can be distributed or due to some other factor(s), such as regulatory hurdles, is unavailable, it is not accessible for use and hence will not be used. Unfortunately, the physical accessibility of potato accessions from global collections is scattered with only 11/23 (~50%) of the collections surveyed having distributed material internationally in the period from 2004 to 2006 (Global Strategy for Ex situ Conservation of Potato 2006). While this is the only statistics found on distribution from multiple global potato global genebanks and it predates the International Treaty for Plant Genetic Resources for Food and Agriculture (Carputo et al. 2013), it serves as a good example and is likely the status today that less than 50% of potato ex situ material held in genebanks globally are available today beyond the walls of the genebank or the borders of the nation in which the genebank resides. This is a reflection of the state of the art of many factors and the international regulatory environment is just one factor (reviewed above). We have also discussed the need for phytosanitary cleaning and a propagule which can be distributed as disease-free. TPS could be an asset and a valuable tool for genebanks lacking the technology for long-term maintenance in tissue culture, yet is capable of virus screening of mother plants for TPS regeneration.
Another major limitation to the use/distribution of potato germplasm is the lack of information publicly accessible about potato germplasm in the various genebanks. While Genesys (https://www.genesys-pgr.org/welcome) offers a one site shop for information on the location of 19,066 potato accessions comprising over 175 species from 21 different countries, as of the end of 2017 it does not have information on 3 (INIA, France; Vavilov, Russia; and NIAS, Japan) out of 5 of the largest potato genebanks identified in the Second Report on The State of the World’s Plant Genetic Resources for Food and Agriculture (FAO 2010). This highlights the difficulty for users to find information on the accessions conserved. Even the European Union Potato Project Database (http://ecpgr.cgn.wur.nl/eupotato/) does not contain information on the French or United Kingdom potato collections, yet it has information from both CIP and USA collections. If information on the holdings of the global potato germplasm collections is not readily available, or if available but not in a universal language, it will be impossible for most users to find information on what is held in ex situ collections.
As mentioned above, one measure of the value of genetic resources conservation is whether the collection is desired by the user community. Using the in trust potato collection at CIP as an example of the value of conservation of diversity of ex situ collections to the breeding and research community, since 2007, CIP has received 1,399 requests for potato germplasm. A total of 48,533 samples were distributed in the last 10 years (2007–2017), following the legal framework of the ITPGRFA. This represents over 1,200 unique accessions per year or almost 25% of the in-trust collection distributed annually. Material was sent to 96 countries with Peru, USA, Australia, and China requesting the most potato germplasm. All material is transferred using the Standard Material Transfer Agreement (SMTA) under the ITPGRFA. It could be expected that in the genomics era, DNA distributions would steadily increase; however, there is no trend in this as physical genetic material remains in high demand suggesting use in breeding programs still requires plants for crossing. Interestingly, in a recent (2015) survey (Shirey, unpublished data), late blight resistance and yield were the most cited traits of interest among users.
In 1997, a program was initiated to return traditional cultivars and landraces to the native, indigenous farmers in Peru (repatriation ), whose ancestors preserved the potato diversity. For centuries, these potato farming communities have nurtured and planted multiple, 20–40, varieties per family as an insurance policy to ensure food availability. The rationale was that in one year one group of landraces would produce well, while in other years a different group might flourish. By conserving and planting diversity, they sustained their livelihood. In recent times, many communities have lost important cultivars they had planted for generations, due to terrorism, extreme weather, climate change, and/or disease/pest pressures. The CIP repatriation program is an example of the broader user base of ex situ genebanks where the in-trust collection served as a reservoir specifically for the landraces the current generation of potato guardians of the Andes (Papa Arariwa) needed to revive their traditional farming systems. In some cases, it allowed them to plant again the landraces they remembered as a child. Thus, repatriation is helping to restore the diversity and productivity in the traditional Andean potato farming region (Aguilar 2016). Further, the repatriation of genebank materials has benefits beyond restoring diversity and the traditions that come with this diversity, the added benefit is the distribution of pathogen-free stocks, which can increase productivity in a single season by as much as 40% thereby contributing to food security and poverty reduction for these communities. In total, 89 communities in Peru have received over 6,000 samples representing more than 1250 accessions of native landraces or over 50% of Peruvian landraces held in the in-trust potato collection.
5 Characterizing Potato Diversity in Genebanks
Molecular markers, are considered fixed landmarks in a genome , and thus, can reveal crucial genetic variability (Semagn et al. 2006). Markers have been used in many crop plants to assess genetic diversity, determine population structure, establishing trait-marker associations, discover and track quantitative trait loci (QTLs), produce genetic linkage maps, assist in selection for traits, understand the influence of genotypes on phenotypes, and more, all to improve or understand crop plants. Loss of plant species in the past centuries has triggered genetic resource conservation with a need to accurately identify each accession. Molecular tools provide an easy less laborious method to assign plant taxa, as well as, characterize certain traits (Arif et al. 2010). To address the challenge of defining the biodiversity of a crop such as potato, biochemical and molecular markers have been used, yet the wide-scale use for looking at entire ex situ collections has only recently become more feasible due to advances in next generation sequencing (NGS) which has made genotyping projects more automated and affordable for large sample sizes.
Examples of where molecular markers have been employed to gain insight in potato are included below. Markers such as isozymes, simple sequence repeats (SSRs), and rapid amplified polymorphic DNA (RAPDs) have been employed in potato for varietal identification and genetic diversity assessments (Anoumaa et al. 2017; Carputo et al. 2013; Collares et al. 2004; Ghislain et al. 2009; Hoque et al. 2013; Rocha et al. 2010; Salimi et al. 2016; Xiaoyan et al. 2016). SSR markers have also been used to support a reevaluation of the taxonomic classification and structure of the gene pool of cultivated potato into four species (Spooner et al. 2007). Amplified fragment length polymorphisms (AFLPs) were utilized to hypothesize the domestication origin for cultivated potato from the northern species of the S. brevicaule group (Spooner et al. 2005) which was contrary to previous reports.
Even though different types of molecular markers exist and have been employed since the 1980s, single nucleotide polymorphisms (SNPs) are increasingly used predominantly due to recent advances in genome sequencing technology, the abundance of SNPs in most crop plants, reduced labor required to collect the data, and price per data point. The affordable cost and high-throughput nature of SNP markers have made them powerful tools for genetic analysis of plant species such as potato and highly useful in breeding (Bertioli et al. 2014). Discovery of SNPs in simple genomes is relatively easy requiring collection and evaluation of sequence data; however, in complex genomes such as potato, SNP detection is more challenging due to repetitive segments of the genome and multiple ploidy levels (Mammadov et al. 2012). Genome complexity reduction methods, such as genotyping by sequencing (GBS), diversity arrays technology (DArTseq), restriction site-associated DNA sequencing (RADseq), have been developed to aid in the discovery of novel SNPs; nevertheless, it is often challenging to identify SNP markers in polyploids such as potato, cotton, canola, and wheat (Bertioli et al. 2014; Logan-Young et al. 2015; Mammadov et al. 2012) due to separating allelic versus homoeologous SNPs or determining dosage in autopolyploids, both of which increase the rate of false positives (Clevenger and Ozias-Akin 2015).
SNP arrays have been developed which allow thousands to one million genome-wide SNP markers to be assessed simultaneously in an individual assay (LaFramboise 2009), thereby reducing the cost per marker data point. The Infinium 12K V2 Potato Array contains 12,720 SNPs, including the SNPs from the original SolCAP Infinium V1 8303 Potato Array with additional markers derived from the Infinium High Confidence SNPs (69K, Hamilton et al. 2011), which were selected for improved genome coverage, candidate genes, and regions with resistance genes. Both potato SNP arrays have been used in numerous studies as a genomic tool to improve cultivated potato or gain insight on genetic attributes. The SolCAP Infinium 8303 Potato Array were selected from 69,011 high-quality SNPs derived from six commercial potato cultivars “Atlantic,” “Premier Russet,” “Snowden,” “Bintje,” “Kennebec,” and “Shepody” (Hamilton et al. 2011). These SNP markers were used to measure linkage disequilibrium for genome-wide association (GWA) mapping and population structure in European diploid and tetraploid germplasm (Stich et al. 2013). Genotyping a diversity panel of 250 lines of wild species, genetic stocks, and cultivated potato revealed that changes in heterozygosity and allele dosage has not occurred in over 150 years of breeding, but clear selection for alleles in biosynthetic pathways has occurred (Hirsch et al. 2013). The SolCAP Infinium 8303 Potato Array was used to develop linkage maps (Felcher et al. 2012), genotype populations for QTL analysis (Douches et al. 2014), and assess variation in glycoalkaloid biosynthesis (Manrique-Carpintero et al. 2013, 2014). It was also used to evaluate the genetic diversity and population structure of S. tuberosum sbsp. andigenum and S. phureja accessions from Colombia along with identifying 23 markers associated with nine morphological traits (Berdugo-Cely et al. 2017). In a wild species study, relationships deduced from the SNP markers were generally complementary to existing taxonomic classifications for 74 Solanum lines representing 25 wild taxa and were also effective in resolving complex taxa boundaries among germplasm with close genetic relationships (Hardigan et al. 2015).
Sequencing efforts including expressed sequence tags (ESTs) and the reference genome for potato (Potato Genome Sequencing Consortium 2011) allowed the development of the Infinium SOLCAP array that has provided the underpinning and a valuable tool for looking more closely at potato diversity. Additionally, the ease of data collection utilizing a SNP array has allowed genotyping of large sample numbers to finally be a reality. The CIP genebank has begun a project to genotype the entire landrace in vitro potato collection utilizing the Infinium V2 12K SNP array (http://solcap.msu.edu/potato_infinium.shtml) along with low density genotyping by sequencing (GBS) of 600 wild potato accessions and over 450 cultivated potatoes. This data is forthcoming; however, it will provide unique fingerprints of a large portion of the collection for users, diversity assessment in wild and cultivated material, identification of potential duplicated material, and genetic tools for genome-wide association studies (GWAS).
An initial assessment of the Infinium V2 SNP array was made by genotyping 250 potato accessions representing 7 taxa (Ellis et al. 2018). The genome-wide SNPs on the array were well distributed across the 12 potato chromosomes ranging from 798 to 1647 SNPs per chromosome. Of the 12,720 SNPs included in the Infinium 12K V2 Potato Array, the majority yielded good-quality signal intensities that were converted into genotypes. SNPs that did not produce a signal in ≥10% of the individuals or could not be clustered were filtered out of the dataset along with SNPs noted in previous studies to be poor or questionable (http://solcap.msu.edu/potato_infinium.shtml). The three-cluster diploid calling yielded 77% (9800) of the total SNPs on the array for use in subsequent analysis. In this data set only, 2.7% of the SNPs were monomorphic with only one of the three diploid genotypic classes scored among all the accessions of the diverse taxa used in this study. Aside from these few, most of the SNPs (97.3%) were polymorphic. A previous study with the 8303-array showed similar rates with a polymorphic rate of 75% (Stich et al. 2013); yet, this study included less taxa with only 44 genotypes. As expected, the five-cluster tetraploid calling had a lower rate of monomorphic markers with only 31 of 4859 (0.6%) of the SNPs being monomorphic; however only 39.5% of the total SNPs on the array could be utilized because of the difficulty in clustering/scoring the three heterozygous classes. Heterozygosity in this data set was high in many species/genotypes and ranged from 0.1 to 81.2% (Ellis et al. 2018).
A distance matrix was calculated from the SNP data and a phylogeny was constructed along with estimates on population structure (Ellis et al. 2018). Most of the taxa did not form distinct monophyletic clades and there were mixed taxa in each clade suggesting significant gene flow, low representation of certain taxa, and/or misclassified accessions. Further, branch lengths among individuals were short suggesting low diversity between taxa and the possibility of hybrids. S. xjuzpeczukii (2n = 3x = 36) was the only taxa to form a distinct monophyletic clade. The accessions of S. xjuzpeczukii were genetically redundant. This is interesting as these accessions originated from three countries (Bolivia, Peru, and Argentina); therefore, one would expect some genetic divergence. However, given that triploids are sterile and thus represent an evolutionary dead-end they were likely spread by human migration as clones. More of the genome needs to be evaluated (i.e., reduced representation sequencing) in these triploid accessions to elucidate if genetic differences among these accessions truly exists or if they are genetic duplicates before an archival process begins of putatively redundant material.
The phylogeny further demonstrated that S. xchaucha, S. stenotomum subsp. goniocalyx, and S. stenotomum subsp. stenotomum appear to be sister taxa with ancestry to S. phureja. Similar suggestions have emerged in previous studies using microsatellite data (Gomez, personal communication) and are supported by the revised taxonomy of Spooner et al. (2014) where these four taxa are lumped into a single species S. tuberosum. The phylogenetic tree also reveals putative discrepancies in species designation of accessions which will need to be verified in the future with phenotyping data. For example, multiple S. stenotomum subsp. stenotomum (17) and S. xchaucha (9) accessions grouped with the S. tuberosum subsp. andigenum accessions. In contrast, most of the S. stenotomum subsp. goniocalyx accessions clustered together or within the S. stenotomum subsp. stenotomum group (Ellis et al. 2018).
The analysis of the SNP data on 250 accessions from the genebank provided valuable information on intra- and interspecific relationships among taxa and provided additional support for targeting the collection of phenotypic data of suspected misclassified accessions. The data suggests that some of the accessions are hybrids and that gene flow has occurred between many of the taxa. The SNP array produced unique fingerprints with a few exceptions among this diverse panel of cultivated potato. These fingerprints provide a legacy for quality management systems (QMS) of the potato collection for years into the future to assure that as accessions are handled errors do not occur, and if they do occur, fingerprints can be used to validate genetic identity. Further, these data can be used to compare accessions between genebanks to determine where unique genetic material exists on a global scale and ensure it is safely preserved for the future. Overall, the genetic analysis facilitated a better understanding of the genetic diversity, population structure, and genetic relatedness of these potato taxa.
6 Climate Change and Genetic Resources Collections
The increased variability of the global climate, resulting in greater extremes of temperatures punctuated by increased dry, wet, cold, warm, and generally unpredictable climatic periods, will continue to have severe impacts on crop productivity and sustainability. Estimates of the extent of temperature increases range between 2 and 6 °C (Jarvis et al. 2011). The warming climates will be accompanied by increased atmospheric CO2, and although both factors will favor plant productivity of C3 plants, such as potatoes, this increased productivity will be accompanied by large increases in biotic (diseases and pests) and abiotic (drought, heat, adverse and unseasonable weather) challenges that could decrease plant yield and increase crop uncertainty. Previously adapted varieties may no longer be robust in the same geographic locality due to a lack of resistance/tolerance to these biotic and abiotic factors. These factors are interrelated, stressed resistance varieties may have attenuated resistance in different environments, resistance may be needed at different developmental stages than previously required, resistance could be overwhelmed by population sizes of pests and diseases, and new pests and diseases may appear. It must also be recognized that one cannot look at a single factor such as how climate affects insect vectors and thus the spread of diseases as an effect on insects will also affect insect pollinators reducing subsequent plant populations, insect predators, and insect populations by an extended growing season. It has been estimated that with a 2 °C rise in temperature, insects will go through one to five additional life cycles/year. In general, bacteria will respond better to moisture fluctuations regardless of temperature while fungi will respond to moisture fluctuations more in cooler weather (Beed 2011).
What is needed now and in the future is genes which can be introgressed into potato to help maintain productivity with the changing climate. Crop wild relatives conserved in genebanks offer “an enormous and unimaginable potential” for the discovery of valuable and desired traits (Machida-Hirano 2015). Wild relatives of potato have been a source of disease resistance for breeding programs for over 100 years (Hawkes 1958). An example is Phytophthora infestans (causal agent of late blight), which is the causal agent responsible for the devastation it inflicted during the Irish potato blight in 1845–1846. It is most severe in periods of high moisture with temperatures between 7.2 and 26.8 °C. It is predicted that for each degree of warming, late blight can occur 4–7 days earlier, extending the period of infections to between 10 and 20 days. With warmer temperatures starting earlier, there is a potential threat that late blight infestation will come earlier and earlier and last longer into the season resulting in increased fungicide applications leading to a greater pressure of the evolution of resistance to the fungicides (Beed 2011). Eleven hypersensitive-type resistance genes (R genes) have been characterized from S. demissum, a wild relative of potato of Mexican origin, thus S. demissum has been used extensively as a source for late blight resistance in European and North American cultivars (Asano and Tamiya 2016; Love 1999). Other sources of resistance in wild relatives of potato include S. bulbocastrum, S. candolleanum, S. chacoense, S. pinnatisectum, and S. stoloniferon (Machida-Hirano 2015).
Warmer temperatures can affect resistance genes rendering them ineffective. In tomato, increased temperatures caused a complete breakdown of resistance to Ralsonia solanacearum (bacterial wilt), another severe disease in potato (Kuun et al. 2001; Tung et al. 1992). Thus, increased temperatures affect not only the pathogenic organisms and vectors but also the resistance genes. Therefore, multiple forms of resistance for breeding programs may be needed and genebanks could provide such multiple forms of resistance. Examples of potato wild relatives where bacterial wilt resistance has been found are S. chacoense, S. commersonii, S. phureja, and S. stonotonum (Asano and Tamiya 2016; Machida-Hirano 2015).
Increased temperatures will also favor an increase in pests including aphids, weevils, and potato tuber moth (Asano and Tamiya 2016), and it has been stated that temperature is likely the single most important factor in cold blooded insect behavior, distribution, development, survival, and reproduction (Bale et al. 2002). Pest resistance in potato wild relatives is associated with high levels of glycoalkaloids, dense hairs, and trichomes found in species such as S. chacoense, S. polyadenium, and S. tarijense (Jansky et al. 2009). Nematode populations can also be influenced by climate as observed with the Southern root-knot nematode (Meloidogyne incognita) in coffee which has expanded its range in recent years associated with changing climatic conditions (Beed 2011). Nematode resistance in potato wild relatives can be found in S. hougasii for Columbia root-knot nematode (Brown et al. 1991) and S. vernei and S. acuale for cyst nematode (Hawkes 1994).
In addition to biotic challenges, climatic changes also can greatly affect potato survival and yield simply by the stresses imposed independent of effects on biotic factors. Increases in early and late season frosts, extreme heat, delay or unseasonable rains and other severe weather events. Tolerance to these abiotic factors is as important as resistance to pests and diseases. S. acuale, a potato wild relative can form a carpet of tiny plants at 4500 m.a.s.l displaying extreme frost and drought tolerance (personal observation). Other frost tolerant species include S. demissum and S. xjuzcepczukii. Drought tolerance can be found in S. chillonanum, S. jamesii, and S. okadae (Watanabe et al. 2011) and S. xcurtilobum (Cabello et al. 2012).
7 Concluding Comments
It is fortunate that the vast majority of the diversity of cultivated potato, as well as much of that of its wild relatives, is already safely conserved in numerous genebanks around the world. These resources represent an investment to ensure potato’s role in food security well into the future. These conserved potato genetic resources will provide the traits and alleles needed to continue potato’s dominance on plates around the world. Increased knowledge of the molecular and biochemical pathways is critical to maximize not only the discovery but also the use of the needed attributes in breeding programs and food systems. The benefits of these ex situ collections will continue to be delivered as the hidden characteristics, some with unknown value today, continue to be uncovered. One cannot however sit back complacently because as long as there is diversity which is threatened, and not securely conserved, there will be valuable alleles, individuals, and populations that could be critical in the future.
References
Aguilar AM (2016) Informe final de siembra de papas nativas repatriadas en la comunidad capesina de Santa Cruz de Pichiu, Chana, Huari, Ancash, 51p
Anoumaa M, Yao NK, Kouam EB, Kanmegne G, Machuka E, Osama SK, Nzuki I, Kamga YB, Fonkou T, Omokolo DN (2017) Genetic diversity and core collection for potato (Solanum tuberosum L.) cultivars from Cameroon as revealed by SSR markers. Am J Potato Res 94:449–463
Arif IA, Bakir MA, Khan HA, Al Farhan AH, Al Homaidan AA, Bahkali AH, Al Sadoon M, Shobrak M (2010) A brief review of molecular techniquest to assess plant diversity. Int J Mol Sci 11:2079–2096
Asano K, Tamiya S (2016) Breeding of pest and disease resistant potato cultivars in Japan using classical and molecular approaches. Jpn Agric Res Q 50(1):1–6
Bajaj YPS (1977) Initiation of shoots and callus from potato-tuber sprouts and axillary buds frozen at −196 °C. Crop Improv 4:48–53
Bale JS, Masters GJ, Hodkinson ID, Awmack C, bezemer TM, Brown VK, Bitterfield J, Buse A, Coulson JC, Farrer J, Good JEG, Harrington R, Hartley S, Jones TM, Lindroth RL, Press MC, Symrnooudis I, Watt AD, Whittaker JB (2002) Herbivory in global climate change research: direct effects of rising temperatures on insect herbivores. Glob Chang Biol 8:1–16
Bamberg J, del Rio A (2011) Use of native potatoes for research and breeding. Hortscience 46(11):1444–1445
Bamberg J, del Rio A, Moreyra R (2009) Genetic consequences of clonal versus seed sampling in model populations of two wild potato species indigenous to the USA. Am J Potato Res 86:367–372
Bamberg J, del Rio A, Fernandez C, Salas A, Vega S, Zorrilla C, Roca W, Tay D (2010) Comparison of “remote” versus “easy” in situ collection locations for USA wild solanum (potato) germplasm. Am J Potato Res 87:277–284. https://doi.org/10.1007/s12230-010-9133-8
Bamberg JB, del Rio A, Huamán Z, Vega S, Salas A, Pavek J, Fernandez C, Spooner DM (2003) A decade of collecting and research on wild potatoes of the Southwest USA. Am J Potato Res 80:159–172
Beed F (2011) The impact of climate change on interdependence for microbial genetic resources for agriculture. In: Fujisaka S, Williams D, Halewood M (eds) The impact of climate change on countries interdependence on genetic resources for food and agriculture. Background Study Paper No. 48. FAO Commission on Genetic Resources for Food and Agriculture, Italy, pp 38–47
Berdugo-Cely J, Valbuena RI, Sanchez-Betancourt E, Barrero LS, Yockteng R (2017) Genetic diversity and association mapping in the Colombian central collection of Solanum tuberosum L. Andigenum group using SNP markers. PLoS One 12(3):e0173039. https://doi.org/10.1371/journal.pone.0173039
Bertioli DJ, Ozias-Akins P, Chu Y, Dantas KM, Santos SP, Gouvea E, Guimaraes PM, Leal-Bertioli SC, Knapp SJ, Moretzsohn MC (2014) The use of SNP markers for linkage mapping in diploid and tetraploid peanuts. G3 (Bethesda) 4:89–96
Bosch FVD, Jeger MJ, Gilligan CA (2007) Disease control and its selection for damaging plant virus strains in vegetatively propagated staple food crops; a theoretical assessment. Proc R Soc [Biol] 247:11–18
Brown CR, Mojtahedi H, Santo GS (1991) Resistance to Columbia root-knot nematode in Solanum spp. and in hybrids of S. hougasii with tetraploid cultivated potato. Am Potato J 68:445–452
Cabello R, De Mendiburu F, Bonierbale M, Monneveux P, Roca W, Chujoy E (2012) Large-scale evaluation of potato improved varieties, genetic stocks and landraces for drought tolerance. Am J Potato Res 89:400–410
Carputo D, Alioto D, Aversano R, Garramone R, Miraglia V, Villano C, Frusciante L (2013) Genetic diversity among potato species as revealed by phenotypic resistances and SSR markers. Plant Genetic Resources 11(2): 131–139. https://doi.org/10.1017/S1479262112000500
Castañeda-Álvarez NP, de Haan S, Juárez H, Khoury CK, Achicanoy HA, Sosa CC, Bernau V, Salas A, Heider B, Simon R, Maxted N, Spooner D (2015) Ex situ conservation priorities for the wild relatives of potato (Solanum L. Section Petota). PLos One 10(14):e0122599
Clevenger JP, Ozias-Akin P (2015) SWEEP: a tool for filtering high quality SNPs in polyploid crops. G3 (Bethesda) 5:1797–1803
Collares EAS, Choer E, Pereira AS (2004) Characterization of potato genotypes using molecular markers. Pesqui Agropecu Bras 39:871–878
Correll DS (1962) The potato and its wild relatives. Contributions from the Texas Research Foundation 4, p 606. Texas Research Foundation, Renner, Texas
De Bruyn A, Villemot J, Lefeuvre P, Villar E, Hoareau M, Harimalala M, Abdoul-Karime AL, Abdou-Chakour C, Reynaud B, Harkins GW, Varsani A, Martin DP, Lett J-M (2012) East African cassava mosaic-like viruses from Africa to Indian Ocean islands: molecular diversity, evolutionary history and geographical dissemination of a bipartite begomovirus. BMC Evol Biol 228(12):228–246
del Rio AH, Bamberg JB, Huaman Z, Salas A, Vega SE (1997) Assessing changes in the genetic diversity of potato gene banks. 2. In situ vs ex situ. Theor Appl Genet 95:199–204
del Rio AH, Bamberg JB, Huaman Z, Salas A, Vega SE (2001) Association of ecogeographical variables and RAPD marker variation in wild potato populations of the USA. Crop Sci 41:870–878
Douches D, Hirsch CN, Manrique-Carpintero NC, Massa AN, Coombs J, Hardigan M, Bisognin D, De Jong W, Buell CR (2014) The contribution of the Solanaceae coordinated agricultural project to potato breeding. Potato Res 57:215–224
Ellis D, Chavez O, Coombs J, Soto J, Gomez R, Douches D, Panta A, Silvestre R, Anglin NL (2018) Genetic identity in genebanks: application of the SolCap 12K SNP array in fingerprinting the global in trust potato collection. Genome 61:523–537
FAO (2010) The second report on the state of the world’s plant genetic resources for food and agriculture. FAO, Rome, 368p
FAO (ed) (2014) Genebank standards for plant genetic resources for food and agriculture, rev edn. FAO, Rome
FAO/IPGRI Technical Guidelines for the Safe Movement of Germplasm. https://cropgenebank.sgrp.cgiar.org/images/file/learning_space/potato_tech_guid_safe_move_germplasm.pdf
Felcher KJ, Coombs JJ, Massa AN, Hansey CN, Hamilton JP, Veilleux RE, Buell CR, Douches DS (2012) Integration of two diploid potato linkage maps with the potato genome sequence. PLoS One 12:e36347
Fuentes XC (2014) Conserving the genetic diversity of Bolivian wild potatoes. PhD thesis, Wageningen University, Wageningen, NL, 229p, ISBN 978-94-6257-168-67
Ghislain M, Nunez J, Herrera MDR, Pignataro J, Guzman F, Bonierbale M, Spooner DM (2009) Robust and highly informative microsatellite based genetic identity kit for potato. Mol Breed 23:377–388
Global Strategy for Ex situ Conservation of Potato 2006. https://www.genebanks.org/wp-content/uploads/2017/01/Potato-Strategy-2007.pdf
Grobman A, Bonavia D, Dillehay TD, Pipernod DR, Iriartef J, Holst I (2012) Preceramic maize from Paredones and Huaca Prieta, Peru. Proc Natl Acad Sci U S A 109(5):1755–1759
Grout BWW, Henshaw GG (1978) Freeze preservation of potato shoot-tip cultures. Ann Bot 42:1227–1229
Hamilton JP, Hansey CN, Whitty BR, Stoffel K, Massa AN, Deynze AV, De Jong WS, Douches DS, Buell CR (2011) Single nucleotide polymorphism discovery in elite North American potato germplasm. BMC Genomics 12:302
Hardigan MA, Bamberg J, Buell CR, Douches DS (2015) Taxonomy and genetic differentiation among wild and cultivated germplasm of Solanum sect Petota. Plant Genome 1:16–270
Hawkes JG (1958) Significance of wild species and primitive forms for potato breeding. Euphytica 7:257–270
Hawkes JG (1989) Nomenclatural and taxonomic notes on the infrageneric taxa of the tuber-bearing solanums (Solanaceae). Taxon 39(3):489–492
Hawkes JG (1990) The potato: evolution, biodiversity and genetic resources. Belhaven Press, London, 259p
Hawkes JG (1994) Origins of cultivated potatoes and species relationships. In: Bradshaw JE, MacKay GR (eds) Potato genetics. CAB International, Wallingford, pp 3–42
Hijmans RJ, Spooner DM (2001) Geographic distribution of wild potato species. Am J Bot 88(11):2101–2112
Hirsch CN, Hirsch CD, Felcher K, Coombs J, Zarka D, Deynze AV, Jong WD, Veilleux RE, Jansky S, Bethke P, Douches DS, Buell CR (2013) Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 (Bethesda) 3:1003–1013. https://doi.org/10.1534/g3.113.005595.
Hoque ME, Huq H, Moon NJ (2013) Molecular diversity analysis in potato (Solanum tuberosum L.) through RAPD markers. SAARC J Agric 11:95–102
International Potato Center (1973–1974) Germplasm exploration and taxonomy of potatoes. In: Program planning conferences 1973–74, Lima, Perú, pp 5–48
International Potato Center (1976) Report of the planning conferences on the exploration and maintenance of germplasm resources, Lima, Peru, Marzo 15–19, 1976, 130p
International Potato Center (1979) Report of the planning conferences on the exploration, taxonomy and maintenance of potato germplasm III. CIP, Lima, Peru, 193p
Jansky SH, Simon R, Spooner DM (2009) A test of taxonomic predictability: resistance to Colorado potato beetle in wild relatives of potato. J Econ Entomol 102:442–431
Jansky SH, Dempewolf H, Camadro EL, Simon R, Zimnoch-Guzowska E, Bisognin DA, Bonierbale M (2013) A case for crop wild relative preservation and use in potato. Crop Sci 53:746–754
Jarvis A, Ramirez J, Hansen J, Leibing C (2011) Crop and forage genetic resources: international interdependence in the face of climate change. In: Fujisaka S, Williams D, Halewood M (eds) The impact of climate change on countries interdependence on genetic resources for food and agriculture. Background Study Paper No. 48. FAO Commission on Genetic Resources for Food and Agriculture, Italy, pp 7–83
Jones RAC (1982) Tests for transmission of four potato viruses through potato true seed. Ann Appl Biol 100:315–320
Kaczmarczyk A, Rokka V-M, Keller ERJ (2011) Potato shoot tip cryopreservation. A review. Potato Res 54:45–79. https://doi.org/10.1007/s11540-010-9169-7
Khan AJ, Akhtar S, Al-Matrushi AM, Briddon RW (2012) Introduction of East African cassava mosaic Zanzibar virus to Oman harks back to “Zanzibar, the capital of Oman”. Virus Genes 46(1):195–198. https://doi.org/10.1007/s11262-012-0838-2
Kinder DH, Adams KR, Wilson HJ (2017) Solanum Jamesii: evidence for cultivation of wild potato tubers by ancestral Puebloan groups. J Ethnobiol 37(2):218–240
Kreuze J (2014) siRNA deep sequencing and assembly: piecing together viral infections. In: Gullino ML, Bonants PJM (eds) Detection and diagnostics of plant pathogens, plant pathology in the 21st century, vol 5. Springer, Dordrecht, 200p. https://doi.org/10.1007/978-94-017-9020-8_2
Kuun GK, Okole B, Bornmann L (2001) Protection of phenypropanoid metabolism by prior heat treatment in Lycopersicon esculentum exposed to Ralstonia solanacearum. Plant Physiol Biochem 39:871–880
LaFramboise T (2009) Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res 37:4181–4193
Legg JP, Jeremiah SC, Obiero HM, Maruthi MN, Ndyetabula I, Okao-Okuja G, Bouwmeester H, Bigirimana S, Tata-Hangy W, Gashaka G, Mkamilo G, Alicai T, Kumar LP (2011) Comparing the regional epidemiology of the cassava mosaic and cassava brown streak virus pandemics in Africa. Virus Res 159(2):161–170
Logan-Young CJ, Yu JZ, Verma SK, Percy RG, Pepper AE (2015) SNP discovery in complex allotetraploid genomes (Gossypium spp., Malvaceae) using genotyping by sequencing. Appl Plant Sci 3:1400077. https://doi.org/10.1007/s12229-014-9146-y
Loskutov IG (1999) Vavilov and his institute. A history of the world collection of plant genetic resources in Russia. International Plant Genetic Resources Institute, Rome. ISBN 92-9043-412-0
Louderback LA, Pavlik BM (2017) Starch granule evidence for the earliest potato use in North America. Proc Natl Acad Sci U S A 114(29):7606–7610
Love SL (1999) Founding clones, major contributing ancestors, and exotic progenitors of prominent North American potato cultivars. Am J Potato Res 76:263–272
Machida-Hirano R (2015) Diversity in potato genetic resources. Breed Sci 65:26–40
Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S (2012) SNP markers and their impact on plant breeding. Int J Plant Genomics 2012:728398. https://doi.org/10.1155/2012/728398
Manrique-Carpintero NC, Tokuhisa JG, Ginzberg I, Holliday JA, Veilleux RE (2013) Sequence diversity in coding regions of candidate genes in the glycoalkaloid biosynthetic pathway of wild potato species. G3 (Bethesda) 3:1467–1479
Manrique-Carpintero NC, Tokuhisa JG, Ginzberg I, Veilleux RE (2014) Allelic variation in genes contributing to glycoalkaloid biosynthesis in a diploid interspecific population of potato. Theor Appl Genet 127:391–405
Ochoa CM (1962) Los Solanun Tuberiferos silvestres del Peru Secc. Tubernarium, sub-secc. Hyperbasarthrum), Lima, Peru
Ochoa CM (1980a) New taxa of Solanum from Peru and Bolivia. Phytologia 46(4):223–225
Ochoa CM (1980b) New tuber-bearing Solanum from Colombia. Phytologia 46(7):495–497. ISSN 0031-9430
Ochoa CM (1980c) Solanum peloquinianum: a new wild potato species from the Peruvian Andes. Am Potato J 57(1):33–35
Ochoa CM (1981a) [Solanum chilliasense, a new tuber bearing species of the Series Piurana]. Solanum chilliasense, nueva especie tuberifera de la serie Piurana. Lorentzia (Argentina) 4:9–11
Ochoa CM (1981b) [Solanum taulisense, a new Peruvian tuber bearing species]. Solanum taulisense, nueva especie tuberifera peruana. Lorentzia (Argentina). 4:13–15
Ochoa CM (1981c) Solanum irosinum, new Peruvian tuber bearing species resistant to Phytophthora infestans. Am Potato J 58(3):131–133. ISSN 0003-0589
Ochoa CM (1981d) Two new tuber-bearing Solanum from South America. Phytologia 48:229–232
Ochoa CM (1983a) Solanum bombycinum, a new tuber-bearing tetraploid species from Bolivia. Am Potato J 60(11):849–852. ISSN 0003-0589
Ochoa CM (1983b) Solanum neovavilovii: a new wild potato species from Bolivia. Am Potato J 60(12):919–923. ISSN 0003-0589
Ochoa CM (1987) Solanum longiusculus (Sect. petota), nova specie peruviana. Phytologia 63(5):329–330. ISSN 0031-9430
Ochoa CM (1988a) [Solanum sarasarae (Sect. Petota), a new species from Perú]. Solanum sarasarae (Sect. Petota), nova specie peruviana. Phytologia 64(4):245–246
Ochoa CM (1988b) Solanum bill-hookeri: new wild potato species from Peru. Am Potato J 65(12):737–740
Ochoa CM (1989a) [Solanum Ser. Simplicissima, new serie Tuberifera of Sect. Petota (Solanaceae)]. Solanum Ser. Simplicissima, nueva serie Tuberifera de la Sect. Petota (Solanaceae). Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales 17(65):321–323. ISSN 0370-3908
Ochoa CM (1989b) Solanum amayanum: a new wild Peruvian potato species. Am Potato J 66(1):1–4. ISSN 0003-0589
Ochoa CM (1989c) Solanum salasianum: new wild tuber-bearing species from Peru. Am Potato J 66(4):235–238. ISSN 0003-0589
Ochoa CM (1990) The potatoes of South America: Bolivia. Cambridge University Press, Cambridge, 512p. ISBN 0-521-38024-3
Ochoa CM (1998) Solanum ortegae: a new Peruvian species from Sect. petota. Phytologia 85(4):271–272. ISSN 0031-9430
Ochoa CM (1999) Las papas de Sudamérica: Perú (parte 1). Allen Press, Kansas, 1036p. ISBN 92-9060-197-3
Ochoa CM (2004) The potatoes of South America: Peru The wild species, part 1. International Potato Center, Peru, 1036p. ISBN 9290601981
Perez W, Salas A, Raymundo R, Huaman Z, Nelson R, Bonierbale M (2001) Evaluation of wild potato species for resistance to Late Blight. In: Scientist and farmer, partners in research for the 21st century, Program Report International Potato Center, Lima, Peru, 480p
Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
Reed BM (2008) Cryopreservation—practical considerations. In: Reed BM (ed) Plant cryopreservation a practical guide. Springer Science, New York, 513p
Ristaino JB, Pfister DH (2016) “What a painfully interesting Subject”: Charles Darwin’s studies of potato late blight. BioScience 66(12):1035–1045
Rocha EA, Paiva LV, de Carvalho HH, Guimaraes CT (2010) Molecular characterization and genetic diversity of potato cultivars using SSR and RAPD markers. Crop Breed Appl Biotechnol 10:204–210
Rumold CU, Aldenderfer MS (2016) Late archaic–early formative period microbotanical evidencefor potato at Jiskairumoko in the Titicaca Basin of southern Peru. Proc Natl Acad Sci U S A 13(48):13672–13677
Ryoko M-H (2015) Diversity in potato genetic resources. Breed Sci 65:26–40
Salimi H, bahar M, Mirlohi A, Talebi M (2016) Assessment of the genetic diversity among potato cultivars from different geographical áreas using the genomic and EST microsatellites. Iran J Biotechnol 14:270–277
Di Santo M, Tarozzi N, Nadalini N, Borini A. (2012) Human sperm cryopreservation: Update on techniques, effect on DNA integrity, and implications for ART. Adv Urol 2012:854837. https://doi.org/10.1155/2012/854837.
Sastry KS, Zitter TA (2014) Plant virus and viroid diseases in the tropics, volume 2: epidemiology and management. Springer, Dordrecht, pp 149–480. https://doi.org/10.1007/978-94-007-7820-7_2
Semagn K, Bjornstad A, Ndjiondjop MN (2006) An overview of molecular marker methods for plants. Afr J Biotechnol 5:2540–2568
Spooner DM, Salas Lopez A, Huaman Z, Hijmans RJ (1999) Wild potato collecting expedition in southern Peru (Department of Apurimac, Arequipa, Cusco, Moquegua, Puno, Tacna) in 1998: taxonomy and new genetic resources. Am J Potato Res 76(3):103–119. ISSN 1099-209X
Spooner DM, Salas AR, Huaman Z, Torres Maita RV, Hoekstra R, Schuler K, Hijmans RJ (2001) Taxonomy and new collections of wild potato species in central and southern Perú. Am J Potato Res 78(3):197–208. ISSN 1099-209X
Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ (2005) A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc Natl Acad Sci U S A 102:14694–14699
Spooner DM, Nunez J, Trujillo G, Herrera MDR, Guzman F, Ghislain M (2007) Extensive simple sequence repeat genotyping of potato landraces supports a major reevaluation of their gene pool structure and classification. Proc Natl Acad Sci U S A 104:19398–19403
Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T (2014) Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot Rev 80:283–383. https://doi.org/10.1007/s12229-014-9146-y.
Stich B, Urbany C, Hoffmann P, Gebhardt C (2013) Population structure and linkage disequilibrium in diploid and tetraploid potato revealed by genome wide high density genotyping using the SolCap SNP array. Plant Breed 132:718–724
Towill LE (1981a) Solanum etuberosum—a model for studying the cryobiology of shoot-tips in the tuberbearing Solanum species. Plant Sci Lett 20:315–324
Towill LE (1981b) Survival at low temperatures of shoot-tips from cultivars of Solanum tuberosum group Tuberosum. CryoLett 2:373–382
Tung PX, Hermsen JG, Zaag P, Vander S, Schmiedche P (1992) Effects of heat tolerance on expression of resistance to Pseudomonas solanacearum in potato. Potato Res 35:321–328
Valkonen JPT, Pehu E, Watanabe K (1992) Symptom expression and seed transmission of alfalfa mosaic virus and potato yellowing virus (SB-22) in Solanum brevidens and Setuberosum. Potato Res 35:403–410
Volk GM, Wadell J, Bonnart R, Towill L, Ellis D, Lauffman M (2008) High viability of dormant Malus buds after 10 years of storage in liquid nitrogen vapour. CryoLetters 29:89–94
Vollmer R, Villagaray R, Egúsquiza V, Cardenas JE, Castro M, Chavez O, Anglin NA, Ellis D (2017) A large-scale viability assessment of the potato cryobank at the International Potato Center (CIP). In Vitro Cell Dev Biol Plant 53(4):309–317. https://doi.org/10.1007/s11627-017-9846-1
Watanabe KN, Kikuchi A, Shimazaki T, Asahima M (2011) Salt and drought stress tolerance in transgenic potatoes and wild species. Potato Res 54:319–324
Wilkinson K, Grant WP, Green LE, Hunter S, Jeger MJ, Lowe P, Medley GF, Mills P, Philipson J, Poppy GM, Waage J (2011) Infectious diseases of animals and plants: an interdisciplinary approach. Philos Trans R Soc B Biol Sci 366(1573):1933–1942
Xiaoyan S, Chunzhi Z, Ying L, Shuangshuang F, Qing Y, Sanwen H (2016) SSR analysis of genetic diversity among 192 diploid potato cultivars. Hortic Plant J 2:163–171
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Ellis, D., Salas, A., Chavez, O., Gomez, R., Anglin, N. (2020). Ex Situ Conservation of Potato [Solanum Section Petota (Solanaceae)] Genetic Resources in Genebanks. In: Campos, H., Ortiz, O. (eds) The Potato Crop. Springer, Cham. https://doi.org/10.1007/978-3-030-28683-5_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-28683-5_4
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-28682-8
Online ISBN: 978-3-030-28683-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)