Abstract
Reprogramming of cells enables generation of pluripotent stem cells and resulting progeny through directed differentiation, making this technology an invaluable tool for the study of human development and disease. Reprogramming occurs with a wide range of efficiency, a culmination of intrinsic and extrinsic factors including the tissue of origin, the passage number and culture history of the target cells. Another major factor affecting reprogramming is the methodology used and the quality of the reprogramming process itself, including for conventional viral-based approaches viral titer and subsequent viral transduction efficiency, including downstream transgene insertion and stoichiometry. Genetic background is an important parameter affecting the efficiency of the reprogramming process with reports that cells from individuals harboring specific mutations are more difficult to reprogram than control counterparts.
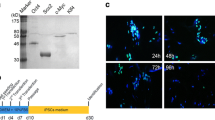
Ataxia-Telangiectasia (A-T) fibroblasts underwent reprogramming at reduced efficiency in contrast to their controls. To optimize reprogramming of fibroblasts from patients with A-T, we examined the response of A-T cells to various cell culture conditions after lentiviral transduction with reprogramming factors Oc4/Sox2 (pSIN4-EF2-O2S) and Klf4/c-Myc (pSIN4-CMV-K2M). Parameters included media type (KSR or serum-containing DMEM), treatment with a p53 inhibitor (small-molecule cyclic pifithrin-α), and either a low or high concentration of bFGF. Post-transduction, equivalent numbers of cells from heterozygote and homozygote patients were plated and assessed at regular intervals for survival and proliferation. Our findings indicate that A-T cells responded favorably to the addition of FCS and gradual weaning away from their native media into KSR-containing stem cell media that produced suitable conditions for their reprogramming. We examined a range of properties to identify and isolate good quality iPSCs including the expression status of important stem cell transcription factors/surface proteins, methylation levels at stem cell associated regulatory loci, persistence of transgenes, karyotype status, and teratoma-forming ability.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Eggan K (2013) Picking the lock on pluripotency. N Engl J Med 369(22):2150–2151
Sommer CA et al (2009) Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27(3):543–549
Plath K, Lowry WE (2011) Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet 12(4):253–265
Lavin MF (2013) The appropriateness of the mouse model for ataxia-telangiectasia: neurological defects but no neurodegeneration. DNA Repair (Amst) 12(8):612–619
Hawley RS, Friend SH (1996) Strange bedfellows in even stranger places: the role of ATM in meiotic cells, lymphocytes, tumors, and its functional links to p53. Genes Dev 10(19):2383–2388
Wood LM et al (2011) A novel role for ATM in regulating proteasome-mediated protein degradation through suppression of the ISG15 conjugation pathway. PLoS One 6(1):e16422
Ambrose M, Goldstine JV, Gatti RA (2007) Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum Mol Genet 16(18):2154–2164
Bar RS et al (1978) Extreme insulin resistance in ataxia telangiectasia: defect in affinity of insulin receptors. N Engl J Med 298(21):1164–1171
Li J et al (2009) Cytoplasmic ATM in neurons modulates synaptic function. Curr Biol 19(24):2091–2096
Lim DS et al (1998) ATM binds to beta-adaptin in cytoplasmic vesicles. Proc Natl Acad Sci U S A 95(17):10146–10151
Cosentino C, Grieco D, Costanzo V (2011) ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J 30(3):546–555
Li J et al (2012) Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med 18(5):783–790
Guo Z, Deshpande R, Paull TT (2010) ATM activation in the presence of oxidative stress. Cell Cycle 9(24):4805–4811
Guo Z et al (2011) ATM activation by oxidative stress. Science 330(6003):517–521
Oka A, Takashima S (1998) Expression of the ataxia-telangiectasia gene (ATM) product in human cerebellar neurons during development. Neurosci Lett 252(3):195–198
Barlow C et al (2000) ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc Natl Acad Sci U S A 97(2):871–876
Song H, Chung SK, Xu Y (2010) Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell 6(1):80–89
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676
Marion RM et al (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460(7259):1149–1153
Kawamura T et al (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460(7259):1140–1144
Polo JM et al (2012) A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151(7):1617–1632
Schubert R, Reichenbach J, Zielen S (2005) Growth factor deficiency in patients with ataxia telangiectasia. Clin Exp Immunol 140(3):517–519
Nayler S et al (2012) Induced pluripotent stem cells from ataxia-telangiectasia recapitulate the cellular phenotype. Stem Cells Transl Med 1(7):523–535
Lin L et al (2015) Spontaneous ATM gene reversion in A-T iPSC to produce an isogenic cell line. Stem Cell Rep 5(6):1097–1108
Fukawatase Y et al (2014) Ataxia telangiectasia derived iPS cells show preserved x-ray sensitivity and decreased chromosomal instability. Sci Rep 4:5421
Lee P et al (2013) SMRT compounds abrogate cellular phenotypes of ataxia telangiectasia in neural derivatives of patient-specific hiPSCs. Nat Commun 4:1824
Maherali N et al (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1(1):55–70
Nishino K et al (2010) DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet 7(5):e1002085
Zhou W et al (2013) Higher methylation in genomic DNA indicates incomplete reprogramming in induced pluripotent stem cells. Cell Reprogram 15(1):92–99
Kim K et al (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467(7313):285–290
Reich M et al (2006) GenePattern 2.0. Nat Genet 38(5):500–501
Chan EM et al (2009) Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol 27(11):1033–1037
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Nayler, S., Kozlov, S.V., Lavin, M.F., Wolvetang, E. (2017). Lentiviral Reprogramming of A-T Patient Fibroblasts to Induced Pluripotent Stem Cells. In: Kozlov, S. (eds) ATM Kinase. Methods in Molecular Biology, vol 1599. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6955-5_29
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6955-5_29
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6953-1
Online ISBN: 978-1-4939-6955-5
eBook Packages: Springer Protocols