Abstract
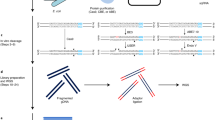
Digenome-seq is a powerful approach for determining the genome-wide specificity of programmable nuclease including CRISPR-Cas9 and CRISPR-Cpf1 (also known as Cas12a) and programmable deaminase including cytosine base editors (CBEs) and adenine base editors (ABEs). To define the genome-wide specificity of dLbCpf1-BE (also known as dLbCas12a-BE), genomic DNA is first incubated with dLbCpf1-BE, which induces C-to-U conversion at on-target and off-target sites, and then treated with a mixture of E. coli uracil DNA glycosylase (UDG) and Endonuclease VIII, which creates single-strand breaks (SSBs) by removing uracil in vitro. Digested genomic DNA is subjected to WGS, and then sequencing reads are aligned to the reference genome, resulting in straight alignments at on-target and off-target sites. The in vitro cleavage sites related to the straight alignments can be identified using the Digenome-seq computer tool.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533(7603):420–424. https://doi.org/10.1038/nature17946
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353(6305). https://doi.org/10.1126/science.aaf8729
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551(7681):464–471. https://doi.org/10.1038/nature24644
Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, Lu Z, Zhang Y, Wu J, Huang X, Yang L, Chen J (2018) Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol 36(4):324–327. https://doi.org/10.1038/nbt.4102
Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, Doudna JA, Liu DR (2020) Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol 38(7):883–891. https://doi.org/10.1038/s41587-020-0453-z
Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, Kim JS (2018) Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 36(6):536–539. https://doi.org/10.1038/nbt.4148
Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, Simon A, Han T, Goswami S, Montgomery E, Thibado J, Kastenhuber ER, Sanchez-Rivera FJ, Shi J, Vakoc CR, Lowe SW, Tschaharganeh DF, Dow LE (2018) Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol 36(9):888–893. https://doi.org/10.1038/nbt.4194
Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR (2018) In vivo base editing of post-mitotic sensory cells. Nat Commun 9(1):2184. https://doi.org/10.1038/s41467-018-04580-3
Kim D, Lim K, Kim DE, Kim JS (2020) Genome-wide specificity of dCpf1 cytidine base editors. Nat Commun 11(1):4072. https://doi.org/10.1038/s41467-020-17889-9
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Kim, D. (2023). Profiling Genome-Wide Specificity of dCpf1 Cytidine Base Editors Using Digenome-Seq. In: Bae, S., Song, B. (eds) Base Editors. Methods in Molecular Biology, vol 2606. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2879-9_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2879-9_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2878-2
Online ISBN: 978-1-0716-2879-9
eBook Packages: Springer Protocols