Abstract
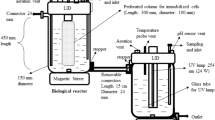
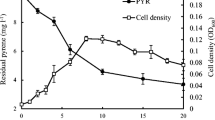
A bacterial strain Pseudomonas sp. a3 capable of degrading nitrobenzene, phenol, aniline, and other aromatics was isolated and characterized. When nitrobenzene was degraded, the release of NH4 + was detected, but not of NO2 −. This result implied that nitrobenzene might have a partial reductive metabolic pathway in strain a3. However, aniline appeared as one of the metabolites during the aerobic degradation of nitrobenzene. Moreover, the appearance of 2-aminophenol during aniline degradation by strain a3 indicated that novel initial reactions existed during the degradation of nitrobenzene and aniline by strain a3. Strain a3 was immobilized in the mixed carrier of polyvinyl alcohol and sodium alginate to improve its degrading efficiency. The optimal concentrations of polyvinyl alcohol and sodium alginate in the mixed carrier were 9 and 3 %, respectively. The immobilized cells had stable degradation activity and good mechanical properties in the recycling tests. The immobilized cells also exhibited higher tolerances in acidic (pH 4–5) and highly saline (10 % NaCl) environments than those of free cells. The biodegradation of nitrobenzene mixed with aniline and phenol using immobilized cells of Pseudomonas sp. a3 was also greatly improved compared with those of free cells. The immobilized cells could completely degrade 300 mg L−1 nitrobenzene within 10 h with 150 mg L−1 aniline and 150 mg L−1 phenol. This result revealed that the immobilized cells of Pseudomonas sp. a3 could be a potential candidate for treating nitrobenzene wastewater mixed with other aromatics.






Similar content being viewed by others
References
Chen D, Chen J, Zhong W, Cheng Z (2007) Degradation of methyl tert-butyl ether by gel immobilized Methylibium petroleiphilum PM1. Bioresour Technol 99:4702–4708
El-Naas MH, Al-Muhtaseb SA, Makhlouf S (2009) Biodegradation of phenol by Pseudomonas putida immobilized in polyvinyl alcohol (PVA) gel. J Hazard Mater 164:720–725
Ha J, Engler CR, Wild JR (2009) Biodegradation of coumaphos, chlorferon, and diethylthiophosphate using bacteria immobilized in Ca-alginate gel beads. Bioresour Technol 100:1138–1142
He Z, Spain JC (1999) Comparison of the downstream pathways for degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45 (2-aminophenol pathway) and by Comamonas sp. JS765 (catechol pathway). Arch Microbiol 171:309–316
Holt GS, Kriey RN, Sneath APH, Staley TJ, Williams TS (1998) Bergey’s manual of determinative bacteriology. Williams and Wilkins, New York
Kaushik D, Pranab R (2011) Degradation of chloroform by immobilized cells of Bacillus sp. in calcium alginate beads. Biotechnol Lett 33:1101–1105
Kulkarni M, Chaudhari A (2007) Microbial remediation of nitro-aromatic compounds: an overview. J Environ Manage 85:496–512
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lane JD (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow E (eds) Nucleic acid techniques in bacterial systematics. Wiley, England
Lessner DJ, Johnson GR, Parales RE, Spain JC, Gibson DT (2002) Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl Environ Microbiol 68:634–641
Liu H, Yang Z, Huang P, Zhou SZ (2002) Degradation of aniline by newly isolated, extremely aniline-tolerant Delftia sp. AN3. Appl Microbiol Biotechnol 58:679–682
Matatov-Meytal YI, Sheintuch M (2002) Hydrotreating processes for catalyticabatement of water pollutants. Catal Today 75:63–67
Miller AS, Dykes DD, Polesky FH (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Mu Y, Yu HQ, Zheng JC, Zhang SJ, Sheng GP (2004) Reductive degradation of nitrobenzene in aqueous solution by zero-valent iron. Chemosphere 54:789–794
Nishino SF, Spain JC (1993) Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl Environ Microbiol 59:2520–2525
Ou LT, Sharma A (1989) Degradation of methyl parathion by a mixed bacterial culture and a Bacillus sp. isolated from different soils. J Agric Food Chem 37:1515–1518
Park HS, Kim HS (2000) Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J Bacteriol 182:573–580
SEPA (2007) Standard methods for the examination of water and wastewater, 4th edn. State Environmental Protection Administration, Beijing
Sergio AMD, Bustos TY (2009) Biodegradation of wastewater pollutants by activated sludge encapsulated inside calcium-alginate beads in a tubular packed bed reactor. Biodegradation 20:709–715
Takenaka S, Okugawa S, Kadowaki M, Murakami S, Aoki K (2003) The metabolic pathway of 4-aminophenol in Burkholderia sp. strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl Environ Microbiol 69:5410–5413
Tao XQ, Lu GN, Liu JP, Li T, Yang LN (2009) Rapid degradation of phenanthrene by using Sphingomonas sp. GY2B immobilized in calcium alginate gel beads. Int J Environ Res Public Health 6:2470–2480
Wang J, Yang H, Lu H, Zhou JT, Zheng CL (2009) Aerobic biodegradation of nitrobenzene by a defined microbial consortium immobilized in polyurethane foam. World J Microbiol Biotechnol 25:875–881
Zheng CL, Qu BC, Wang J, Zhou JT, Wang J, Lu H (2009a) Isolation and characterization of a novel nitrobenzene-degrading bacterium with high salinity tolerance: Micrococcus luteus. J Hazard Mater 165:1152–1158
Zheng CL, Zhou JT, Wang J, Qu BC, Lu H, Zhao HX (2009b) Aerobic degradation of nitrobenzene by immobilization of Rhodotorula mucilaginosa in polyurethane foam. J Hazard Mater 168:298–303
Zheng CL, Zhou JT, Wang J, Qu BC, Wang J, Lu H, Zhao HX (2009c) Aerobic degradation of 2-picolinic acid by a nitrobenzene-assimilating strain: Streptomyces sp. Z2. Bioresour Technol 100:2082–2084
Acknowledgments
This work was supported by Chinese National Natural Science Fund (31070099, 30830001 and 31000060), the Opening Fund of State Key Laboratory of Soil and Sustainable Agriculture (Y052010025), the Fundamental Research Funds for the Central Universities (KYZ200920) and the Major Technology Special Social Development Project (2007C13059).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, Z., Liu, Y., Liu, H. et al. Characterization of the nitrobenzene-degrading strain Pseudomonas sp. a3 and use of its immobilized cells in the treatment of mixed aromatics wastewater. World J Microbiol Biotechnol 28, 2679–2687 (2012). https://doi.org/10.1007/s11274-012-1078-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-012-1078-2