Abstract
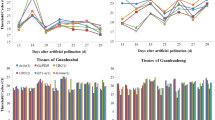
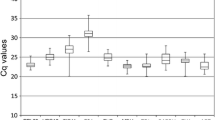
Reverse transcription quantitative real-time PCR (RT-qPCR), a sensitive technique for quantifying gene expression, depends on the stability of the reference gene(s) used for data normalization. Only a few studies on reference genes have been done in fruit trees and none in litchi. In the present study, seven frequently used candidate reference genes, including actin (ACTIN), glyceraldehyde-3-phosphate-dehydrogenase (GADPH), elongation factor 1-alpha (EF-1α), poly ubiquitin enzyme (UBQ), α-tubulin (TUA), β-tubulin (TUB) and RNA polymerase-II transcription factor (RPII), were evaluated for their expression stability in litchi. A total of 78 samples, including different varieties, tissues, organs, developmental stages and treatments, such as NAA, shading and girdling plus defoliation, were addressed in this analysis. Our results showed that GAPDH was the most suitable reference gene among all the tested samples, different organs and NAA treatment. ACTIN was stably expressed in varieties and fruit developmental stages. RPII and UBQ exhibited better expression stability in tissues. EF-1α was the most stable gene in shading and girdling plus defoliation treatments. Moreover, using combination of two genes as reference genes might improve the reliability of gene expression by RT-qPCR in litchi. A better combination was GAPDH + EF-1α or GAPDH + ACTIN for all the examined samples. In addition, the validated reference genes were further relied on to quantify the expression of an interested gene, LcARF13 under different experimental conditions. These results first provide guidelines for reference genes selection under different experimental conditions and also a foundation for more accurate and widespread use of RT-qPCR in litchi.





Similar content being viewed by others
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10:49
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Mol Endocrinol 29:23–29
Bustin SA, Dorudi S (1998) Molecular assessment of tumour stage and disease recurrence using PCR-based assays. Mol Med Today 4:389–396
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Chyau CC, Ko PT, Chang CH, Mau JL (2003) Free and glycosidically bound aroma compounds in lychee (Litchi chinensis Sonn.). Food Chem 80:387–392
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LMG, Romano E, Grossi-de-Sa M, Vaslin M, Alves-Ferreira M (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breed 23:607–616
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? J Exp Bot 55:1445–1454
Guilfoyle TJ, Ulmasov T, Hagen G (1998) The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell Mol Life Sci 54:619–627
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol 6:609–618
Hibbeler S, Scharsack JP, Becker S (2008) Housekeeping genes for quantitative expression studies in the three-spined stickleback Gasterosteus aculeatus. BMC Mol Biol 9:18
Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8:112
Huang XM, Suranant S, Mitra SK, Ben-Arie Stern R (2005) Origin, history, production and processing. In: Menzel C, Waite GK (eds) Litchi and Longan: botany, production and uses. CABI Publishing, Oxford
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization strategies and considerations. Genes Immun 6:279–284
Iskandar HM, Simpson RS, Casu RE, Bonnett GD, Maclean DJ, Manners JM (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Rep 22:325–337
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophy Res Commun 345:646–651
Jiang YM (2000) Role of anthocyanins, polyphenol oxidase and phenols in lychee pericarp browning. Sci Food Agric 80:305–310
Jiang YM, Li YB (2003) Effects of low-temperature acclimation on browning of litchi fruit in relation to shelf life. J Hortic Sci Biotech 78:437–440
Jiang YM, Duan XW, Joyce D, Zhang ZQ, Li JR (2004) Advances in understanding of enzymatic browning in harvested litchi fruit. Food Chem 88:443–446
Li JG, Huang HB, Gao FF, Huang XM, Wang HC (2001) An overview of litchi fruit cracking. Acta Hort 558:205–208
Li QF, Sun SSM, Yuan DY, Yu HX, Gu MH, Liu QQ (2010) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57
Lin YL, Lai ZX (2010) Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci 178:359–365
Lovdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242
Lu WJ, Jiang YM (2003) Clone sequence analysis of two enlarged gene in Litchi fruit. Sci Agr Sin 6:1525–1529 (in Chinese with English abstract)
Lu WJ, Wang Y, Jiang YM, Li JG, Liu H, Duan XW, Song L (2006) Differential expression of litchi XET genes in relation to fruit growth. Plant Physiol Biochem 44:707–713
Maroufi A, Van Bockstaele E, De Loose M (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15
Martin RC, Hollenbeck VG, Dombrowski JE (2008) Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Sci 48:1881–1887
Munish A, Kahlon PS, Mahajan BVC (2003) Effect of exogenous application of growth regulators on fruit drop, cracking and quality of litchi (Litchi chinensis Sonn.) Cv. Dehradun. Agr Sci Dig 23:191–194
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Silveira ED, Alves-Ferreira M, Guimaraes LA, da Silva FR, Carneiro VTD (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9:84
Terzi V, Morcia C, Spini M, Tudisco R, Cutrignelli MI, Infascelli F, Stanca AM, Faccioli P (2010) Identification and validation of reference genes for gene expression studies in water buffalo. Animal 4:853–860
Tong ZG, Gao ZH, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:71
Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C (2002) Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem 309:293–300
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, DePaepe A, Speleman F (2002) Accurate normalisation of realtime quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1–research0034.11
Wan HJ, Zhao ZG, Qian CT, Sui Y, Malik AA, Chen JF (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Wang Y, Lu WJ, Li JG, Jiang YM (2006) Differential expression of two expansin genes in developing fruit of cracking-susceptible and -resistant litchi cultivars. J Am Soc Hortic Sci 131:118–121
Wu SX (1998) Encyclopaedia of China fruits: Litchi. China Forestry Press, Beijing
Xiang X, Zhang YS, Huang SZ, Ou Lx, Qiu YP, Li ZQ, Chen JZ (2007) Application of suppressive subtraction hybridization in the study of embryo abortion related to gene expression in litchi (Litchi chinensis Sonn.). Acta Hortic 763:83–89
Yang YF, Hou S, Cui GH, Chen SL, Wei JH, Huang LQ (2010) Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol Biol Rep 7:507–513
Yi C, Jiang YM, Shi J, Qu HX, Xue S, Duan XW, Shi JY, Prasad NK (2010) ATP-regulation of antioxidant properties and phenolics in litchi fruit during browning and pathogen infection process. Food Chem 118:42–47
Zampieri M, Ciccarone F, Guastafierro T, Bacalini MG, Calabrese R, Moreno M, Villanueva A, Reale M, Chevanne A, Bürkle P, Caiafa (2010) Validation of suitable internal control genes for expression studies in aging. Mech Age D 131:89–95
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (30871694), Doctoral Fund of Ministry of Education of China (200805640003) and Modern Agro-industry Technology Research System (grant No. nycytx-32-03).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Lakshmanan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhong, HY., Chen, JW., Li, CQ. et al. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30, 641–653 (2011). https://doi.org/10.1007/s00299-010-0992-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0992-8