Abstract
The recent outbreak of the respiratory pandemic known as novel coronavirus SARS-CoV-2 disease “COVID-19” was first identified in Wuhan, China and quickly spread to other countries. The 3CL protease (3CLpro) enzyme is the main protease of the SARS-CoV-2, which is responsible for coronavirus replication and therefore, the 3CLpro is considered a drug discovery target. The study reports that the molecular docking approach of 30 compounds had been identified from Allium roseum against two 3CL proteases (3CLpro) targets (PDB: 6LU7) (PDB: 6M2N) of SARS-CoV-2 using Autodock Vina. The docking results revealed that the top three compounds, Kaempferol-7-O-rutinoside, Kaempferol-3-O-glucuronoside and Apigenin-7-O-glucoside, displayed high affinity against the two 3CLproteas binding pockets. The free energy of binding (FEB) were − 12.10, − 11.80 and − 11.52 against 6LU7 and − 11.83, − 11.34 and − 11.01 kcal/mol against 6M2N for Autodock, while AutoDock Vina scores were − 11.6, − 11.2 and − 10.3 kcal/mol against 6LU7 and − 11.1, − 10.8 and − 10.5 kcal/mol against 6M2N. The results reveal that the three compounds fully interact with the essential amino acids in the binding pocket catalytic dyad Cys145 and His41 of the two 3CLpro. Consequently, they are expected to hinder SARS-CoV-2 3CLpro activity. In conclusion, our In-silico results suggest that the three identified compounds could serve as a potential lead that could inhibit the function of 3CLprotease (3CL pro) of Coronavirus. However, in-vitro and in-vivo experiments are necessary to certify and confirm the docking results reported here.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The Coronavirus 2019 (Covid19) outbreak of severe acute respiratory syndrome (SARS) has spread worldwide, with a growing number of victims. According to WHO data on the November 6, 2020, the Coronavirus had infected 219 countries, resulting in 47,596,852 cases and 1,216,357 deaths [1]. The rapid transmission from human to human has increased the number of victims day by day, even though no special antiviral medications have been given to the afflicted patients [2]. Different approved FAD drugs such as anti‐influenza (oseltamivir) [3], HIV antiviral agents (lopinavir) [4], and antimalarial (chloroquine) [5] have been used in the treatment of Coronavirus. It helped a little bit with the urgent condition, however, they may still be trial and error because no selective medicine has been discovered yet. Infection of the upper respiratory tract is caused by Coronaviruses (CoVs) in higher mammals and humans [6]. Several outbreaks have been linked in the recent past to CoVs. Originally identified as SARS in 2002, MERS in 2012, and COVID-19 in late 2019 [7,8,9,10] and [11]. The coronavirus SARS-CoV-2 is a positive-sense, an enveloped, single-stranded RNA virus that belongs to the family Coronaviridae [12]. Its phylogenetically similar to the Middle East respiratory syndrome (MERS) and the severe acute respiratory syndrome coronavirus (SARS) coronavirus [13]. Coronavirus bio-macromolecules are important functional biological macromolecules. The viral protease (3CLpro) and RNA-dependent RNA-polymerase (RdRp) are required for polyprotein proteolytic processing and viral replication. Therefore, they have been proposed as potential therapeutic targets in treating viral diseases [14]. Inhibiting the activity of the 3CLpro enzyme consider as one of the most characterized therapeutic targets for coronaviruses. Furthermore, it is important to treat the polyproteins translated from RNA molecules [15, 16]. The 3CLpro monomer consists of three domains, namely domain I, domain II and domain III, and a long loop connect domains II and III. In addition, the 3CLpro active site has a Cys-His catalytic dyad (Cys145 and His41) positioned in the gap between domains I and II [8, 9, 17, 18]. SARS-CoV-2 3CLpro active site has a Cys-His catalytic dyad (Cys145 and His41), and it’s found in the gap between domains I and II [19]. The primary objective of the scientific community is to develop effective drugs. This objective needs a systematic limiting of pharmacological candidates, which is typically accomplished by virtual screening techniques [20]. In-silico drug screening employing several computer-aided drug design applications, the rational drug design process is substantially accelerated. Ultimately, this saves time and money because it eliminates the need to experiment with leads that fail during the drug discovery process [21,22,23]. The most promising technique for speeding up the discovery of new drug candidates is in-silico drug discovery. It's becoming increasingly important in drug development, and they are critical in identifying promising therapeutic candidates at a low cost [24]. Many drugs found in the market have been discovered of natural origin. Natural origin can be classified into three groups: unaltered natural (pure) products, a mixture of natural product (NP), and natural product derivatives obtained from plants or other living organisms, such as lichens, fungi, microorganisms, sponges [25]. Traditional medicine is becoming more widely recognized as a useful treatment option for various illnesses, and large funds are being invested in research into the therapeutic potential of medicinal plants. In addition, several phytochemicals with significant antiviral activity have been reported in the literature. These phytochemicals could be used to restrict coronavirus replication [26]. The Alliaceae family includes multiple genera, with the Allium genus being the most widely distributed and important, with more than 700 cultivated species. Although the species varies in color, form and taste, they have similar biochemical, phytochemical, and nutraceutical qualities. Furthermore, this genus is economically significant as a source of agricultural and herbal crops and a decorative plant [27, 28]. Allium roseum L. (vernacular name: lazoul) is a polymorphous species endemic in North Africa from western Libya to Marocco [29] from the Liliaceae family, is a rich source of organo-sulfur compounds and flavonoids, it is worth mentioning that this medicinal plant is widely used in north Libya to treat influenza. The Allium roseum plant contains many compounds isolated from different parts of the plant from the flowers, stems, leaves, and bulbs during the plant's growth cycle: the vegetative phase and the flowering phase (February). Many compounds have been isolated from the essential oil using hydrodistillation methods followed by gas chromatography-mass spectroscopy (GC–MS), Another compounds isolated from different plant extracts such as (methanolic extracts, ethanolic extracts, and ethyl acetate extracts) characterized using the high-performance liquid chromatography-mass spectroscopy (HPLC–MS) [30], Liquid Chromatography Electrospray Ionization Mass Spectroscopy Most of the isolated compounds exhibited different biological activities such as antibacterial, antifungal, antiviral, anticarcinogenic, antithrombotic, antiatherosclerotic, immuno-stimulating, and antioxidant properties and cholesterol-lowering effects Allium species [31, 32]. Several compounds have been isolated from the Allium roseum L. from different parts of the plant, such as flavonoids, phenolic and sulphurous compounds (Quinic acid, Protocatechuic acid, Trans ferulic acid, Apigenin-7-O-glucoside, Luteolin-7-O-glucoside, Kæmpferol, Apigenin, Trans ferulic acid, 2,4-dimethylthiophene, 2-propenyl methyl, disulfide, Oxygenated sesquiterpene and p-Cymene) [30, 33,34,35,36].
The present study aimed to use the in-silico molecular docking approach to identify and evaluate potential inhibitors against two SARS-CoV-2 3CL proteases obtained from the Allium roseum L. plant, followed by molecular docking analysis to find new inhibitors that could be utilized as potential leads in the treatment of coronavirus infections.
2 Materials and Methods
2.1 Protein Preparation
The 3D structure of two used proteins, SARS-CoV-2 main protease (Mopar, 3CLpro) (PDB ID: 6LU7) [17,18] in complex with a peptide-like inhibitor N3 and (PDB ID: 6M2N) [37] in complex with Baicalein were obtained from Protein Data Bank (https://www.rcsb.org/). First, the water molecules and co-factor were removed and hydrogen was added using AutoDockTools (ADT) [38].
2.2 Ligand Preparation
The 3D structures of natural product compounds were obtained from the PubChem [39] site in SDF format and then converted to pdb to be used in molecular docking using Discovery studio visualizer [40].
2.3 Molecular Docking
AutoDock Vina [41] and AutoDock 4.2 [42] were used to perform molecular docking. The centre grid parameter was specified as 60–60–60 for x-, y- and z-axes, respectively, a spacing of 0.375 Å and located at the centre of the active site.
3 Results and Discussion
3.1 Validation of the Docking Protocol
The docking protocol has been validated by re-docking the coordinated ligand from the crystallographic structure of COVID-19 main proteinases (PDB ID: 6LU7 and PDB ID: 6M2N) into the same binding pocket. The docking results showed a similarity between the ligand pose and coordinated pose (RMSD = 0.88 Å, 0.97 Å, (Fig. 1), the binding affinity − 7.5 kcal/mol and − 8.2 kcal/mol) for 6LU7 and 6M2N, respectively. The obtained results displayed that the docking protocol used in this study is reliable because the RMSD value was less than the 2.0 threshold value established to evaluate reliability; the result suggests that the docking protocol utilized is accurate [43]. The conserved residues found in the binding pocket of 3CLprotease were targeted in this investigation to block virus activity.
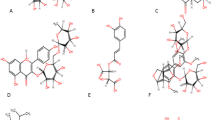
Firstly 30 reported compounds identified from Allium roseum L. were obtained from the literature. Three candidates, Kaempferol-7-O-rutinoside, Kaempferol-3-O-glucuronoside and Apigenin-7-O-glucoside, as shown in (Fig. 2), were chosen based on the lowest FEB of the protein–ligand complex AutoDock Vina.
In the docking study, all the 30 compounds showed low binding energy ranging from − 10.3 to − 0.8 kcal/mol and − 10.5 to − 0.7 kcal/mol by AutoDock Vina for 6LU7, 6M2N, respectively. The protein–ligand complex that exhibited the lowest free energy of binding is considered best [44]. Consequently, the top 3 ranked compounds were suggested as the potential candidates. Kaempferol-7-O-rutinoside, Kaempferol-3-O-glucuronoside and Apigenin-7-O-glucoside displayed lowest FEB − 12.10, − 11.80 and − 11.52 kcal/mol for 6LU7 and − 11.83, − 11.34 and − 11.01 kcal/mol for 6M2N by AutoDock 4.2 and − 11.6, − 11.2 and − 10.3 kcal/mol for 6LU7 and − 11.1, − 10.8 and − 10.5 kcal/mol for 6M2N by AutoDock Vina (Table 1). AutoDock orders the obtained results by FEB and based on the cluster of solutions that utilizes the same pose as shown in (Table 2) [45]. The results of Kaempferol-7-O-rutinoside displayed that the largest poses, 39 poses (consider the largest cluster out of 100 poses), adopted a favourable conformation. On the other hand, the compound Kaempferol-3-O-glucuronoside adopted the pose 22 times out of 100. Apigenin-7-O-glucoside also adopted this pose 27 times out of 100 for 6LU7. The results of compound Kaempferol-7-O-rutinoside displayed that 28 poses. For Kaempferol-3-O-glucuronoside, adopted showed the largest poses among others 42 poses out of 100, while Apigenin-7-O-glucoside adopted this pose 33 times out of 100 for 6M2N shown in (Table 2).
AutoDock reports the optimal docking solution lowest FEB and cluster analysis for each GA run, including the total number of clusters and the rank of each docking mode (cluster rank). The number of clusters for each docked compound, the range of docked energies, the cluster rank of the docking mode and the docked energy of the docking mode chosen is all shown in (Table 2). It is worth mentioning that only the docking mode with the lowest FEB was selected from this cluster for most of the docking.
The essential amino acid residues completely wrapped the three selected compounds at the active binding pocket region (Fig. 3), showed that all the four compounds together with the crystallographic ligands present inside the binding pockets of the two 3CLpro indicate that they are interacted with the essential amino acids, making them compete with the coordinated ligands.
Each of the three compounds’ docking sites was studied. The findings revealed that they all used the same binding region in the protein and had similar strengths suggesting that this site could be attacked and subsequently covalently bound by the essential amino acid residues at the binding pocket. In this study, we manually investigated the results of the docked molecules and their interactions with the target proteins. Then the location of the docked compounds was determined at the binding pocket (Fig. 4), with their structures reaching toward the corresponding component. As a result, the potential compounds formed extensive interactions with the amino acid residues that constructed the binding pocket, as showing in (Fig. 4, 5). The formed interactions include hydrophobic, van deer walls, electrostatics, Pi-Cation, T-sigma, Pi-alkyl and hydrogen bonds. Furthermore, the Interaction results of each compound with both proteins revealed that these three compounds interacted with most of the residues in the binding pocket, as shown in (Fig. 4, 5). For the compounds against 6LU7, Kaempferol-7-O-rutinoside was found to show five hydrogen bonds; the first one formed between the amino acid THR 190 and first oxygen atom O1, another between His163 and second oxygen atom O2, likewise Gly143, Ser144 formed three hydrogen bonds with the third oxygen atom O3. In addition, van der Waals interaction formed between THR26, ASN142, LEU141, MET156, ASP187, TYR45, GLN192 and the carbon atoms C-1, C-2, C-6, C-7, C-9, C-10, C-15, C-16 and C-20 of the compound. Likewise, the amino acid CYS145 exhibited Pi-Sulfur bonds with the first benzene ring on the compounds. Two amino acids MET49, HIS41 displayed Pi-alkyl bonds on the fifth benzene ring. The other interaction was the Carbon hydrogen bond between amino acids HIS4, ARG188, GLN189, CLU166 and Pi-Sigma displayed between HIS41 and C25 on the compound Table 3 and Fig. 5.
The second compound was Kaempferol-3-O-glucuronoside was found to interact with GLU166, GLU166, ASN142, HIS136 through hydrogen bonds with oxygen atoms O6, O8, O9 and O10. Van der walls interactions were noticed between amino acids ASP187, HIS41, TYR54, ARG188, GLN189, MET165, AER144, LEU141, GLY143 and carbon atoms C-1, C-6, C-9, C-10, C-11, C-12, C-13, and C-18 on the compound. Likewise, the Pi-Sulfur bond displayed between CYS145 and the first benzene ring on the compound. Amino Acid MET49 exhibited two Pi-sigma bonds with the second and third benzene rings on the compounds Table 3 and Fig. 5. The third compound was Apigenin-7-O-glucoside was found to show three hydrogen bonds between the amino acids THR190, GLY143, LEU141 and three oxygen atoms on the compound. Van der walls interaction displayed between the amino acids SER144, ASN142, THR25, THR26, HIS41, MET49, GLN189, ARG188, GLN192 and carbon atoms C-1, C-17, C-18, C-19, C-20 and C-21. Likewise, CYS145and MET156 amino acids formed two Pi-Alkyl bonds with first and third benzene rings, respectively Table 3 and Fig. 5. While the three selected against the protein, 6M2N, the first compound was Kaempferol-7-O-rutinoside displayed four hydrogen bonds between the amino acids ARG188, GLU166, GLU166, THR26 and oxygen atoms O1, O6, O7 and O14 on the compounds. The amino acids GLN192, PRO168, LEU167, MET165, GLN198, ASP187, GLY143, LEU27, THR25, CYS44, TYR54, ASP187, GLN178 formed van der Waals interactions with the carbon atoms C-1, C-3, C-9, C-10, C-11, C-12, C-13, C-14, C-15 and C-16 on the compound. Pi-alkyl bonds were formed between amino acids MET49, CYS45 and first and fifth benzene rings. In addition, MET49 formed Pi-Sigma with the second benzene ring. Likewise, a Carbon hydrogen bond was displayed between MET49 and the second benzene ring Table 3 and Fig. 5. The second compound was Kaempferol-3-O-glucuronoside, found to show five hydrogen bonds between GLY143, GLU166, CLU166, asn142, CYS145 with the oxygen atoms O5, O7, O8, O10 and O10. Van der walls interactions formed between amino acids THR25, THR26, LEU27, MET165, GLN189, ARG188, TYR54, ASP187 and C1, C2, C3, C4, C11, C12, C13 and C14. Pi-Sulfur bond was formed between CYS44 and the first benzene ring. MET49 formed two Pi-Sigma bonds with first and second benzene rings. Likewise, the Pi-Cation bond showed between the His41 and second benzene ring Table 3 and Fig. 5. The third compound was Apigenin7-O-glucoside exhibited five hydrogen bonds between amino acids residues GLY143, THR26, HIS41, GLU166 and oxygen atoms O1, O2, O3, O7 and O10. Van der Waals interaction formed between amino acid residues LEU167, PRO168, GLN192, GLN189, ARG188, ASP187, TYR54, CYS44, THR25, LEU27, CYS145 and C1, C2, C3, C4, C5, C7, C9, C11, C12 and C13. MET165 forms a Pi-Sulfur bond with the first benzene ring on the compound. Pi-Cation bond formed with the amino acid HIS41as well as Pi-Sigma and Pi-Alkyl bonds displayed with the MET49 amino acid Table 3 and Fig. 5.
In this study, in-silico structure-based drug design was used to speed up the discovery of SARS-Coronavirus-2 antiviral compounds using a rational and inexpensive approach [46, 47]. According to the above findings, three natural compounds identified from Allium roseum L. displayed high affinity and good binding interactions against the two 3CLpro. Consequently, it can bind to the catalytic dyad Cys145 and His41 simultaneously with other amino acid residues found in the binding pocket. Therefore, these compounds are anticipated to inhibit 3CLpro activity. As a result, we recommended these compounds be potent protease inhibitors for COVID-19. In addition, the results of the four Allium roseum L. compounds against a two 3CLp showed molecular properties, minimum FEB, hydrogen-bond, binding modes, and electrostatic interactions between the essential amino acid residues at the binding pocket and the compounds. Our research included molecular properties screening, two docking approaches, and hydrogen bond analyses. Although the structural analyses and molecular features employed in our simulations were simple, they helped minimize the number of docked compounds and computational costs. In addition, they increased the accuracy and reliability of the results.
Natural products and plant extracts have long been known to have significant antiviral properties, with inhibitory effects on viral enzymes essential to the viral life cycle. Certain natural products inhibit CoV virulence by inhibiting viral proteins such as 3CLpro, PLpro, S, and ACE2, in addition to their inhibitory effect on viral replication or virulence. Due to the genomic similarities and similarity in the main protease structure and the principal host receptor between SARS-CoV and SARS-CoV-2, natural compounds that have shown efficacy against SARS-CoV can be used as a selection criterion for potential inhibition of SARS-CoV-2 [48, 49]. Many classes of natural product compounds were identified from various plants and investigated against the Coronavirus used in in-silico and in-vitro studies, where many of these compounds proved their effectiveness and could potentially inhibit the SARS-COV-1 and SARS-COV-2, such as 7-methoxycryptopleurine, lycorine, silvestrol, tylophorine, homoharringtonine and ouabain [50], epicatechin-gallate, luteolin7-O-glucoside, apigenin-7-O-glucoside, quercetin, catechin, naringenin, oleuropein, curcumin and desmetho-xycurcumin [51] and hesperetin, glycyrrhizin, nicotianamine, scutellarin and baicalin [52]. A study by Jo et al. [53] showed that flavonoids from traditional Chinese medicines like Herbacetin, Rhoifolin and Pectolinarin effectively blocked SARS-CoV 3CLpro enzymatic activity by inhibition of the viral life cycle inside the host of SARS-CoV-2 Mpro,the IC50 values were 33.17, 27.45 and 37.78 µM, for herbacetin, rhoifolin and pectolinarin, respectively. In silico and in vitro studies demonstrated that quercetin had been shown to interfere with many phases of the coronavirus entrance and replication cycle, including PLpro, 3CLpro, and NTPase/helicase [54]. Furthermore, Hesperidin is a flavanone glycoside that was able to Inhibit both ACE2 and virus entrance into the cells host by binding to the helicase, S protein, and protease sites on the ACE receptor [55]. Given the genetic similarities, it’s not surprising that natural product compounds that are effective against SARSCoV1 could also be effective against SARSCoV2. The obtained results from two different studies of the same compounds revealed that these three compounds are potent inhibitors of the SARS-CoV Mpro with IC50 values of 73 µM, 47 µM and 73 µM for quercetin, gallocatechin gallate and pigallocatechin gallate, respectively against SARS-CoV-1 [56] and then in-silico docking and molecular dynamics simulations of the same compounds displayed low binding affinity and different interactions with the essential amino acids in the binding pocket of the target protein PDB: 6LU7 of COV-2 [57]. These results confirm the possibility of using the natural compounds that showed activity against SARS-COV-1 as inhibitors for SARS-COV-2. Comparing our findings to those previously reported studies in terms of the used protein, similar classes of compounds, and the docking simulation methods confirmed the validity of our in-silico investigation conducted in this study. In addition, our results confirmed that the identified three compounds from this plant could potentially inhibit the coronavirus SARS-COV-2.
4 Conclusion
In-silico molecular docking and interaction analyses were successfully applied to identify natural compounds as lead candidates identified from Allium roseum L. against two 3CL proteases (3CLpro) target SARS-CoV-2. The three identified compounds, Kaempferol-7-O-rutinoside, Kaempferol-3-O-glucuronoside and Apigenin-7-O-glucoside, displayed a high binding affinity with the 3CLpro binding pocket of COVID-19. The free energy of binding (FEB) were − 12.10, − 11.80 and − 11.52 against 6LU7 and − 11.83, − 11.34 and − 11.01 kcal/mol against 6M2N for Autodock, while AutoDock Vina scores were − 11.6, − 11.2 and − 10.3 kcal/mol against 6LU7 and − 11.1, − 10.8 and − 10.5 kcal/mol against 6M2N. The obtained results exhibited that the interactions between the compounds and the conserved catalytic dyad Cys145 and His41 and other amino acids were similar to the coordinated ligands N3 and baicalein 6LU7 6M2N, respectively. Experimental studies in-vitro and in-vivo research is needed to transform these potential inhibitors into therapeutic drugs. Nevertheless, we anticipate that the findings of this work will be useful in the future for investigating and synthesized novel natural anti-COVID-19 therapeutic agents.
References
World Health Organization (2020) World Health Organization official website. Accessed 6 Nov 2020. https://www.who.int/emergencies/diseases/novelcoronavirus-2019.
Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433.
Letchumanan V, Ab Mutalib NS, Goh BH, Lee LH. Novel coronavirus 2019-nCoV: could this virus become a possible global pandemic. Prog Microbes Mol Biol. 2020. https://doi.org/10.36877/pmmb.a0000068.
Sahin AR, Erdogan A, Agaoglu PM, Dineri Y, Cakirci AY, Senel ME, Okyay RA, Tasdogan AM. 2019 Novel coronavirus (COVID-19) outbreak: a review of the current literature. EJMO. 2020;4(1):1–7.
Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924.
Yang Y, Peng F, Wang R, Guan K, Jiang T, Xu G, Sun J, Chang C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434.
Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, Zumla A. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–6.
Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–88.
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–9.
Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16(10):1686.
Ul Qamar MT, Saleem S, Ashfaq UA, Bari A, Anwar F, Alqahtani S. Epitope-based peptide vaccine design and target site depiction against Middle East Respiratory Syndrome Coronavirus: an immune-informatics study. J Transl Med. 2019;17(1):1–14.
Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–36.
Tobaiqy M, Qashqary M, Al-Dahery S, Mujallad A, Hershan AA, Kamal MA. Therapeutic management of COVID-19 patients: a systematic review. Infect Prev Pract. 2020;2(3):100061.
Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–47.
Ghosh AK, Xi K, Grum-Tokars V, Xu X, Ratia K, Fu W, Houser KV, Baker SC, Johnson ME, Mesecar AD. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg Med Chem Lett. 2007;17(21):5876–80.
Khan RJ, Jha RK, Amera GM, Jain M, Singh E, Pathak A, Singh RP, Muthukumaran J, Singh AK. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J Biomol Struct Dyn. 2021;39(8):2679–92.
Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–93.
Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C. Structure of Mpro from covid-19 virus and discovery of its inhibitors. Nature. 2020;582(7811):289–93.
Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn. 2021;39(7):2673–8.
Alves DR, Da Rocha MN, de Sousa DS, Oliveira ICM, Marinho MM, de Morais SM, Marinho ES. Virtual screening of natural curcumins and related compounds against SARS-CoV-2. J Comput Biophys Chem. 2021;20(01):53–70.
Abdusalam AAA, Murugaiyah V. Identification of potential inhibitors of 3CL protease of SARS-CoV-2 from ZINC database by molecular docking-based virtual screening. Front Mol Biosci. 2020;7:419.
Maia EHB, Assis LC, de Oliveira TA, da Silva AM, Taranto AG. Structure-based virtual screening: from classical to artificial intelligence. Front Chem. 2020;8:343.
Muneer I, Tusleem K, Abdul Rauf S, Hussain HM, Siddiqi AR. Discovery of selective inhibitors for cyclic AMP response element-binding protein: a combined ligand and structure-based resources pipeline. Anticancer Drugs. 2019;30(4):363–73.
Brogi S, Ramalho TC, Kuca K, Medina-Franco JL, Valko M. In silico methods for drug design and discovery. Front Chem. 2020;8:612.
Lautie E, Russo O, Ducrot P, Boutin JA. Unraveling plant natural chemical diversity for drug discovery purposes. Front Pharmacol. 2020;11:397.
Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592.
Snoussi M, Trabelsi N, Dehmeni A, Benzekri R, Bouslama L, Hajlaoui B, Al-sieni A, Papetti A. Phytochemical analysis, antimicrobial and antioxidant activities of Allium roseum var. odoratissimum (Desf.) Coss extracts. Ind Crops Prod. 2016;89:533–42.
Tepe B, Sokmen M, Akpulat HA, Sokmen A. In vitro antioxidant activities of the methanol extracts of five Allium species from Turkey. Food Chem. 2005;92(1):89–92.
Hichem BJ, Lamia SR, Mohamed N, Zine M. Chemical composition of flowers and stems essential oils from the Tunisian Allium roseum L. J Essent Oil Bear Plants. 2007;10(2):151–6.
Boubakri A, Leri M, Bucciantini M, Najjaa H, Ben Arfa A, Stefani M, Neffati M. Allium roseum L. extract inhibits amyloid beta aggregation and toxicity involved in Alzheimer’s disease. PLoS ONE. 2020;15(9):e0223815.
Pittler MH, Ernst E. Clinical effectiveness of garlic (Allium sativum). Mol Nutr Food Res. 2007;51(11):1382–5.
Štajner D, Igić R, Popović BM, Malenčić DJ. Comparative study of antioxidant properties of wild growing and cultivated Allium species. Phytother Res Int J Devot Pharmacol Toxicol Eval Nat Prod Deriv. 2008;22(1):113–7.
Najjaa H, Neffati M, Zouari S, Ammar E. Essential oil composition and antibacterial activity of different extracts of Allium roseum L., a North African endemic species. C R Chim. 2007;10:820–6.
Najjaa H, Zerria K, Fattouch S, Ammar E, Neffati M. Antioxidant and antimicrobial activities of Allium roseum L. “Lazoul”, a wild edible endemic species in North Africa. Int J Food Prop. 2011;14(2):371–80.
Zouari S, Najjaa H, Neffati M, Ammar E. A new essential oil chemotype of Allium roseum analyzed by an apolar column. Int J Food Prop. 2012;15(2):385–97.
Zouari S, Ketata M, Boudhrioua N, Ammar E. Allium roseum L. volatile compounds profile and antioxydant activity for chemotype discrimination–case study of the wild plant of Sfax (Tunisia). Ind Crops Prod. 2013;41:172–8.
Su HX, Yao S, Zhao WF, Li MJ, Liu J, Shang WJ, Xie H, Ke CQ, Hu HC, Gao MN, Yu KQ. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin. 2020;41(9):1167–77.
Ikram NKK, Durrant JD, Muchtaridi M, Zalaludin AS, Purwitasari N, Mohamed N, Rahim ASA, Lam CK, Normi YM, Rahman NA, Amaro RE. A virtual screening approach for identifying plants with anti H5N1 neuraminidase activity. J Chem Inf Model. 2015;55(2):308–16.
Bolton EE, Chen J, Kim S, Han L, He S, Shi W, Simonyan V, Sun Y, Thiessen PA, Wang J, Yu B. PubChem3D: a new resource for scientists. J Cheminformatics. 2011;3(1):1–15.
BIOVIA. Dassault systems, discovery studio visualizer, v16.1.0.15350. San Diego: Dassault Systèmes; 2015.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61.
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–91.
Bourne Y, Taylor P, Radić Z, Marchot P. Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site. EMBO J. 2003;22(1):1–12.
Spyrakis F, Amadasi A, Fornabaio M, Abraham DJ, Mozzarelli A, Kellogg GE, Cozzini P. The consequences of scoring docked ligand conformations using free energy correlations. Eur J Med Chem. 2007;42(7):921–33.
Smith DM, Daniel KG, Wang Z, Guida WC, Chan TH, Dou QP. Docking studies and model development of tea polyphenol proteasome inhibitors: applications to rational drug design. Proteins Struct Funct Bioinform. 2004;54(1):58–70.
Abdusalam AAA, Vikneswaran M. Novel acetylcholinesterase inhibitors identified from ZINC database using docking-based virtual screening for Alzheimer’s disease. ChemistrySelect. 2020;5(12):3593–9.
Hariono M, Rollando R, Karamoy J, Hariyono P, Atmono M, Djohan M, Wiwy W, Nuwarda R, Kurniawan C, Salin N, Wahab H. Bioguided fractionation of local plants against matrix metalloproteinase9 and its cytotoxicity against breast cancer cell models: in silico and in vitro study. Molecules. 2020;25(20):4691.
Chakravarti R, Singh R, Ghosh A, Dey D, Sharma P, Velayutham R, Roy S, Ghosh D. A review on potential of natural products in the management of COVID-19. RSC Adv. 2021;11(27):16711–35.
Verma S, Twilley D, Esmear T, Oosthuizen CB, Reid AM, Nel M, Lall N. Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19). Front Pharmacol. 2020;11:1514.
Islam MT, Sarkar C, El-Kersh DM, Jamaddar S, Uddin SJ, Shilpi JA, Mubarak MS. Natural products and their derivatives against coronavirus: a review of the non-clinical and pre-clinical data. Phytother Res. 2020;34(10):2471–92.
Khaerunnisa S, Kurniawan H, Awaluddin R, Suhartati S, Soetjipto S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints. 2020. https://doi.org/10.20944/preprints202003.0226.v1.
Chen H, Du Q. Potential natural compounds for preventing 2019- nCoV infection. Preprints. 2020. https://doi.org/10.20944/preprints202001.0358.v3.
Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzym Inhib Med Chem. 2020;35(1):145–51.
Agrawal PK, Agrawal C, Blunden G. Quercetin: antiviral significance and possible COVID-19 integrative considerations. Nat Prod Commun. 2020;15(12):1934578X20976293.
Llivisaca-Contreras SA, Naranjo-Morán J, Pino-Acosta A, Pieters L, Vanden Berghe W, Manzano P, Vargas-Pérez J, León-Tamariz F, Cevallos-Cevallos JM. Plants and natural products with activity against various types of coronaviruses: a review with focus on SARS-CoV-2. Molecules. 2021;26(13):4099.
Nguyen TTH, Woo HJ, Kang HK, Nguyen VD, Kim YM, Kim DW, Ahn SA, Xia Y, Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotech Lett. 2012;34(5):831–8.
Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. 2020. https://doi.org/10.1080/07391102.2020.1779818.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there is No Conflict of Interest.
Rights and permissions
About this article
Cite this article
Benhander, G.M., Abdusalam, A.A.A. Identification of Potential Inhibitors of SARS-CoV-2 Main Protease from Allium roseum L. Molecular Docking Study. Chemistry Africa 5, 57–67 (2022). https://doi.org/10.1007/s42250-021-00296-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-021-00296-y