Abstract
Experiments were carried out to study the effect of zinc oxide nanoparticles (nano-ZnO) on nitrogenase activity in legumes. In the first experiment, nodulated roots of cluster bean, moth bean, green gram and cowpea were dipped in Hoagland solution containing 1.5 and 10 μg mL−1 of nano-ZnO for 24 h. Nitrogenase activity in cluster bean, green gram and cowpea roots increased after dipping in solution containing 1.5 μg mL−1 nano-ZnO, but decreased in roots dipped in solution containing 10 μg mL−1 nano-ZnO. However, in moth bean roots, nitrogenase activity decreased after dipping in solution containing either concentration of nano-ZnO. In the second experiment, nodulated roots of green gram were dipped in Hoagland solution containing 1, 4, 6, 8 and 10 μg mL−1 nano-ZnO for 6–30 h before estimating nitrogenase activity. Results showed that an interactive effect of nano-ZnO concentration and exposure time influenced nitrogenase activity. The possible reasons behind this effect have been discussed. A model [A = 3.44 + 0.46t − 0.01t 2 − 0.002tc 2 (R 2 = 0.81)] involving linear and power components was developed to simulate the response of nitrogenase activity in green gram roots to the concentration and exposure time of nano-ZnO.
Similar content being viewed by others
Introduction
About 10,000 tonnes of engineered metal nanoparticles of CeO, CuO, CuI, ZnO, TiO2 and elemental Ag are annually produced in the world for industrial and commercial applications [1, 2]. Large quantities of these nanoparticles eventually end up in agricultural soils and may affect microbial-mediated nutrient transformations processes, e.g. immobilization, denitrification, nitrification and nitrogen fixation which help in sustaining the soil and ecosystem health. Heinlaan et al. [3] reported toxicity of nano-ZnO to Vibrio fischeri. The antimicrobial activity of ZnO, CuO and Fe2O3 nanoparticles against Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis and Staphylococcus aureus has also been reported [4–7]. Nanoparticles of CeO, Ag, Cu and chitosan are also reported to be toxic to nitrifying bacteria (Nitrosomonas and Nitrobacter), E. coli and B. subtilis [7–9]. Vitousek et al. [10] reported that 90–140 TgN is annually fixed through biological nitrogen fixation, which is the second most important process after photosynthesis carried out by plants. Therefore, the adverse effect of nanoparticles on nitrogen fixation can have serious ramifications [11–13]. In this paper, we present the experimental results on the effect of concentration and exposure time of zinc oxide (ZnO) nanoparticles on nitrogenase activity in four legumes.
Materials and methods
Characterization of ZnO nanoparticles
Nanoparticles of ZnO prepared and characterized by the Indian Institute of Technology-Bombay, Mumbai, were used in the present study [14]. ZnO nanoparticles were prepared by reacting Zn(O2CCH3)2 dissolved in methanol with methylated KOH. The purity of the compound was found to be 99.5 %. The shape of the particles was spherical and their size ranged from 16 to 30 nm. The surface area of the synthesized ZnO nanoparticles was 23 m2 g−1 as determined by multipoint Brunauer–Emmett–Teller (BET) method.
Raising plants in pots
Ten seeds of four legumes, viz. cowpea [Vigna unguiculata (L.) Walp.; c.v. RC101], green gram [Vigna radiata (L.) Wilczek: c.v. K-851], clusterbean [Cyamopsis tetragonoloba, (L.) Taub.; c.v. RGC 936] and moth bean [Vigna aconitifolia (Jacq.) Marechal; c.v. RMO-40] were sown in pots filled with 12 kg soil. Soil pH in the 1:2 (soil:water) mixture was 8.1. Sand, silt and clay content of the soil was 85.1, 9.7, 4.5 %, respectively, whereas organic matter content was 1.5 g kg−1. Ten days after germination, the number of plants in each pot was reduced to five. Plants were allowed to grow under natural conditions in a net house till the flowering stage. Thirty pots were maintained for each crop, i.e. cowpea, green gram, cluster bean and moth bean to obtain adequate number of replicates. Additionally, green gram was also grown in 200 other pots. Plants in pots were irrigated periodically. Plants were gently uprooted at the flowering stage. Water was applied before uprooting to avoid root damage. The effect of nano-ZnO on nitrogenase activity in roots was estimated by transferring and maintaining the plants under hydroponic condition as detailed below.
First experiment
Exposing roots to nano-ZnO
Roots of plants uprooted from soil were dipped in glass containers filled with 1 l of Hoagland solution of ¼th strength [15] as shown in Fig. 1. Full-strength Hoagland solution consisted of 20 µg mL−1 (NH4)2SO4, 10 µg mL−1 NH4NO3, 3.1 µg mL−1 NaH2PO4, 40 µg mL−1 K2SO4, 15 µg mL−1 CaCl2·2H2O, 0.35 µg mL−1 EDTA·FeNa·3H2O, 25 µg mL−1 MgSO4·3H2O, 20 µg mL−1 Al2(SO4)3·18H2O, 0.1 µg mL−1 ZnSO4·7H2O, 0.1 µg mL−1 H3BO3, 0.025 µg mL−1 CuSO4·5H2O, 1 µg mL−1 MnSO4·H2O, and 0.05 µg mL−1 Na2MoO4·2H2O and was prepared by using AR-grade chemicals (Fluka/Hi-media). For each crop, a set of five glass containers were used. The first chamber contained ¼th strength of Hoagland solution comprising 1.5 µg mL−1 and the second had 10 µg mL−1 nano-ZnO. Hoagland solution in the third and fourth chamber was mixed with bulk ZnO (Analytical Grade) to give 1.5 and 10 µg mL−1 ZnO concentration, respectively. The fifth chamber was filled with only ¼th strength Hoagland solution. After 24 h, plants were taken out of the chamber, their shoots were cut and roots transferred to incubation bottles for estimating nitrogenase activity as detailed in the subsequent paragraph. Nitrogenase activity in roots dipped in Hoagland solution alone served as control and that in the solution containing bulk ZnO was used for comparison with corresponding nano-ZnO treatments. At the same time, nitrogenase activity in three plant roots of each crop was also estimated soon after their removal from pots (without dipping them in Hoagland solution) with a view to assess the effect of dipping of roots in Hoagland solution on nitrogenase. All analyses were carried out in triplicate.
Estimation of nitrogenase activity
Each of the de-topped plant roots with attached nodules was kept in an incubation bottle fitted with lid containing a self-sealing septum at the centre. Ten mL of air was taken out from the bottle and an equal volume of acetylene was injected. Bottles along with roots were incubated for 1 h at 27 °C and then 1 mL of gas sample was taken out with the help of a Hamilton syringe and immediately fed into GC (Nucon 5675) fitted with flame ionization detector (FID) and 2 m-length Porapak stainless steel column (id 2 mm). Nitrogen was used as carrier gas. The temperature of the column was kept at 70 °C and that of the injection port maintained at 110 °C. The retention time of ethylene was 1.2 min. The amount of ethylene produced by roots was derived after injecting a range of standard ethylene (110–1000 µg mL−1) supplied by Spancan Products Limited, England. After estimation of nitrogenase, the roots were washed, and the nodules were detached and weighed.
Second experiment
Green gram showed a very wide range of response to nano-ZnO application in the first experiment; therefore, the second experiment on the interaction of concentration and exposure duration to nano-ZnO was envisaged only with this crop. Its roots were exposed to 1, 4, 6, 8 and 10 µg mL−1 nano-ZnO for 6, 12, 18, 24 and 30 h, respectively. The other procedural details were akin to those described earlier for the first experiment.
Modelling the effect of concentration and time of exposure to nano-ZnO on nitrogenase activity
The response of green gram to nano-ZnO application with respect to concentration and exposure time was simulated, initially using models of Chick-Watson and Hom [16]. However, as satisfactory fit was not observed, efforts were made to develop a new model structure with the observed data in the general linear form as follows:
where “Y” is the nitrogenase activity, “c” is the concentration of nano-ZnO in Hoagland solution, “t” is the time of incubation, “a” is the intercept of linear model, “b i” is the coefficients of independent variable or input of the model and “x ”i is the independent variable or input of the model, which may be either “t” or “c” or their interactions.
Stepwise procedure was first followed to select significant inputs from an initial set of inputs involving c and t and their interactions up to the third power. Then the linear models were developed with selected inputs from the stepwise procedure. The whole procedure of model development was done in R mathematical software with ‘step’ and ‘lm’ function.
The developed model was validated using leave-out-one cross-validation approach, where randomly (N − 1) observations were selected from the original dataset to develop the model and then tested on the left-out-one observation, which was repeated for N times, where N is the number of observations in the dataset [17]. The observed and predicted values from cross-validation were plotted against the 1:1 line and root mean square error (RMSE) was calculated to show the fitting performance:
where Y i is the observed value of nitrogenase activity, \(\hat{Y}_{i}\) is the predicted value of the same and N is the number of observations in the dataset.
Results and discussion
The effect of nano-ZnO on nitrogenase in nodulating roots of selected legumes has been presented and discussed in the subsequent paragraphs so as to understand the effect of its concentration and exposure duration.
Response to bulk and nano-ZnO in legumes
Transfer of intact plant from soil to hydroponics and subsequent estimation of nitrogenase activity in de-topped roots with attached nodules did not result in appreciable change in all the legumes studied except cluster bean (Table 1). This could be due to a thin water film that remained adhered around cluster bean roots even at the time of nitrogenase assay. Roots of different plants vary in their composition of lignin, celluloses, etc., which imparts to them differing composition and surface characteristics [18, 19]. Thus, it seems that roots of cluster bean provided more conducive surface for adhesion of water film than roots of other legumes studied. The presence of water film around the roots can alter the diffusion of acetylene and ethylene from roots and thereby influence the nitrogenase activity, since the diffusion coefficient in water is about 1000 times less than in air [20].
The presence of 1.5 µg mL−1 bulk ZnO in Hoagland solution decreased nitrogenase activity in all the legumes. The maximum reduction was recorded in moth bean (71.2 %), while the minimum was in green gram (22.9 %) roots. However, a similar concentration of nano-ZnO stimulated nitrogenase activity in all the legumes except moth bean. Stimulation of nitrogenase activity in Nostoc at 0.625 µg g−1 zinc has also been reported by Okmen et al. [21]. The stimulatory effect of hydroxyapatite nanoparticles on pectate lyase up to 10.5 µg mL−1 due to chaperon-like activity has also been reported by Dutta et al. [22]. Additionally, the elevation of native state activity of the enzyme may be responsible for the observed increase as reported for pectate lyase in the presence of hydroxyapatite nanoparticles [23]. Higher stability of keratinase exposed to iron oxide nanoparticles is also known [24]. As for the present investigation on nitrogenase, either one or all of these reasons may be responsible for the stimulatory response to ZnO nanoparticles application up to a certain concentration.
At 10 µg mL−1, both bulk and nano-ZnO were noted to show a declined nitrogenase activity with respect to control in all the legumes studied. Further, nitrogenase in green gram, moth bean and cowpea was more adversely affected by nano-ZnO than the bulk ZnO. However, such a trend was not observed in cluster bean, the reasons for which are not immediately clear. Differences in the effect of bulk and nano-ZnO could be due to better penetration by the latter in the cells which may result in a variety of responses [25–31]. Nanoparticles have the ability to attach to bacterial membrane via electrostatic interaction and disrupt its integrity [32]. Because of their simpler cell membrane structure, Gram-negative bacteria, like all rhizobium strains involved in legume nodulation, are more adversely affected by nanoparticles [33, 34]. The interaction of nanoparticles with cellular organelles, redox active proteins such as NADPH oxidase and cell surface receptors can generate reactive oxygen species (ROS), which may cause oxidative stress and DNA damage, thus disrupting cell division [33, 35–39]. The release of soluble ions which bind with the functional groups of protein resulting in their denaturation has also been associated with the toxic effects of nano-ZnO, nano-CuO and Ag nanoparticles [4, 40]. Thus, it is possible that any or all of these mechanisms may explain the observed reduction of nitrogenase by nano-ZnO.
Interplay of concentration and time
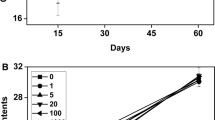
Nitrogenase activity in green gram roots was exposed to 1 µg mL−1 nano-ZnO changed with exposure time. First, it increased with exposure time, reached a maximum at 12 h and then declined (Fig. 2). A similar trend was observed with 4 µg ml−1 nano-ZnO. After 30 h of exposure, nitrogenase activity in roots which were exposed to 1 µg mL−1 nano-ZnO remained only slightly more than in the control. However, in roots exposed to 4 µg mL−1, activity lower than in control was recorded. At 6 and 8 µg mL−1, nano-ZnO nitrogenase activity was maximum at 18 h and the negative effect on increasing exposure time was more pronounced. Exposing roots to 10 µg mL−1 nano-ZnO even for 6 h decreased nitrogenase activity which reduced to zero after 24 h. Thus, both concentrations of nano-ZnO and the exposure time influenced nitrogenase activity. Consequently, three trends were observed: (1) short time exposure of roots to <10 µg mL−1 nano-ZnO enhanced nitrogenase activity (2) increasing concentrations of nano-ZnO adversely affected nitrogenase activity and (3) increasing exposure time also adversely affected nitrogenase activity. Therefore, nitrogenase activity in green gram indicated an interactive effect between concentration of nanoparticles and duration of exposure.
An important factor in understanding the mechanism of nanoparticles-induced changes in nitrogenase is to take into account their interaction with proteins. The dispersion of nanoparticles in a biological milieu results in their surface being immediately enveloped by a complex layer of protein known as “protein corona” [41, 42]. This adsorption of a protein on the surface of nanoparticles strongly depends on its nature, surface chemistry and physicochemical properties. Subsequently, adsorption, electrostatics, hydrogen bonding and hydrophobic interactions provide further binding between protein and nanoparticles. This leads to conformational change in the protein [43]. Thus, the interaction of the nanoparticles with enzymes may be indirect and occurs via nanoparticle–protein corona and not at the bare nanoparticle surface [42]. As nature, size, shape, charge and hydrophobicity of proteins vary depending on plant species and their metabolites, the interactive effect of metabolites with nanoparticles would also be different. These interactions between nanoparticles and protein can acquire both negative and positive connotations.
Chakraborti et al. [44] reported that nano-ZnO induced unfolding of the periplasmic domain in Vibrio cholerae that made its protein susceptible to denaturation. Similarly, Sinha and Khare [45] reported structural changes in protease in the presence of nano-ZnO as the reason behind reduction in its activity. It is possible that structural changes in nitrogenase may also constitute a reason behind the observed decrease in its activity at 10 µg mL−1 nano-ZnO concentration (Table 1).
However, the structural changes can also improve the enzyme stability as in the case of α-helical content of lysozyme in the presence of nano-ZnO [46]. Miletic et al. [47] also reported that immobilization of enzyme on polystyrene nanoparticles resulted in their increased activity. Thus, enhanced stabilization of nitrogenase at low concentrations of nano-ZnO could be one possible mechanism to explain the observed enhanced activity. The second mechanism could be the creation of novel conformational epitomes in nitrogenase during unfolding in the process of its interaction with nanoparticle–protein corona surface. Unfolding may also lead to exposure of hidden epitopes. These new epitopes may affect the functionality of the bound proteins by elicitation of a new response. Such a possibility for different enzymes has been discussed by [48–52]. Further, the direct enzyme–nanoparticle attachment may also facilitate enzyme–substrate interaction by preventing aggregation of free enzyme [22].
Modelling nitrogenase activity
A model for describing the nitrogenase activity (A) as a function of time of incubation (t) and concentration (c) of nano-ZnO formulation has been developed. Initial analysis of data showed that the activity first increased with increasing both t and c up to a peak level and then decreased with further increase in either t or c. Such response was first visualized in 3-D surface using a perspective plot of R, which indicated the response surface as a cubic function (Fig. 3).
Model describing nitrogenase activity as a function of exposure time and nanoparticle concentration
The cubic response surface of nitrogenase activity as depicted in Fig. 3 was tried to fit in the linear model involving linear and interactive components of t and c (t, c, t 2, c 2, tc, t 3, t 2 c, tc 2 and c 3). Stepwise analysis in both backward and forward approach screened three significant variables: t, t 2 and tc 2, and the following model was developed:
All the coefficients and intercept of Eq. (3) were significant (p < 0.001). The intercept of Eq. 3 indicates the enzymatic activity at t = 0 and c = 0. The fitted surface of enzymatic activity as a function of t and c is shown in Fig. 4. The developed model showed that both t and c, indicated as time of incubation and concentration of nano-ZnO, played an important role in enzymatic activity. Modelling the sole effect of either t or c resulted in poor performance in fitting the observed enzymatic activity.
Validation results of the developed model
The model was further checked using leave-out-one cross-validation approach. The observed and fitted values are presented in Fig. 5. The observed and predicted plots showed very close proximity to the 1:1 line with neither overprediction nor underprediction. Moreover, the RMSE of prediction was very low, 0.981 (observed data range of nitrogenase from negligible to 8.27). This indicates satisfactory performance of the developed model, and the same model structure is likely to predict accurately the behaviour of nitrogenase activity under a similar set of experimental conditions for unknown concentrations and exposure durations.
On the basis of these results, it can be concluded that the effect on nano-ZnO on nitrogenase varies with both concentration and exposure time. Low concentration of nano-ZnO enhances nitrogenase activity, but its effect is ephemeral, i.e. enhanced nitrogenase activity is lowered if exposed for a longer duration. Such a response is explained through one or more than one mechanism at the bio-chemical plane as discussed in the preceding paragraphs. Further, a mathematical model, as developed in the present investigation, can satisfactorily simulate the response of nitrogenase to the concentration of nano-ZnO over time. This information could further be exploited to regulate nitrogenase activity, a key process of nitrogen fixation, through use of nanoparticles for enhancing N enrichment of nodulating legumes.
References
Hendren, C.O., Mesnard, X., Dröge, J., Weisner, M.R.: Estimating production data for five engineered nano-materials as a basis for exposure assessment. Environ. Sci. Technol. 45, 2562–2569 (2011)
Piccinno, F., Gottschalk, F., Seeger, S., Nowack, B.: Industrial production quantities and uses of ten engineered nanomaterials in Europe and the world. J. Nanoparticle Res. (2012). doi:10.1007/s11051-012-1109-9
Heinlaan, M., Ivask, A., Blinova, I., Dubourguier, H., Kahru, A.: Toxicity of nano sized and bulk ZnO, CuO and TiO2 to bacteria Vibrio fischeri and crustaceans Daphnia magna and Thamnocephalus platyurus. Chemosphere 71, 1308–1316 (2008)
Azam, A., Ahmed, A.S., Oves, M., Khan, M.S., Habib, S.S., Memic, A.: Antimicrobial activity of metal oxide nanoparticles against gram-positive and gram negative bacteria: a comparative study. Int. J. Nanomed. (2012). doi:10.2147/IJN.S35347
Sotiriou, G.A., Pratsinis, S.E.: Antibacterial activity of nano-silver ions and particles. Environ. Sci. Technol. (2010). doi:10.1021/es101072s
Li, M., Zhu, L., Lin, D.: Toxicity of ZnO nanoparticles to Escherichia coli: mechanism and the influence of medium components. Environ. Sci. Technol. (2011). doi:10.1021/es102624t
Choi, O., Hu, Z.: Size dependent and reactive oxygen species related nano-silver toxicity to nitrifying bacteria. Environ. Sci. Technol. (2008). doi:10.1021/es102624t
Pelletier, D.A., Suresh, A.K., Holton, G.A., McKeown, C.K., Wang, W., Gu, B., Allison, M.R., Brown, S.D., Phelps, T.J., Doktycz, M.J.: Effects of engineered cerium oxide nanoparticles on bacterial growth and viability. Appl. Environ. Microb. 76, 7981–7989 (2010)
Qi, L., Xu, Z., Jiang, X., Hu, C., Zou, X.: Preparation and antibacterial activity of chitosan nanoparticles. Carbohyd. Res. 339, 2693–2700 (2004)
Vitousek, P.M., Menge, D.N., Reed, S.C., Cleveland, C.C.: Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos. Trans. R. Soc. B (2013). doi:10.1098/rstb.2013.0119
Sylvia, D.M., Fuhrmann, J.J., Hartel, P., Zuberer, D.A.: Principles and Applications of Soil Microbiology. Pearson Prentice Hall, New Jersey (2005)
Chapin III, F.S., Matson, P., Pamela, A., Vitousek, P.: Principles of Terrestrial Ecosystem Ecology, 2nd edn. Springer, New York (2012)
Cherchi, C., Gu, A.Z.: Impact of titanium dioxide nanomaterials on nitrogen fixation rate and intracellular nitrogen storage in Anabaena variabilis. Environ. Sci. Technol. 44, 8302–8307 (2010)
Mahajan, P., Dhoke, S.K., Khanna, A.S., Tarafdar, J.C.: Effect of Nano-ZnO on growth of mungbean (Vigna radiata) and chickpea (Cicer arietinum) seedlings using plant agar method. Appl. Biol. Res. 13, 54–61 (2011)
Hoagland, D.R., Arnon, D.I.: The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 347, 1–32 (1950)
Hass, C.N., Kaymak, B.: Effect of initial microbial density on inactivation of Giardia muris by ozone. Water Res. 37, 2980–2988 (2003)
Allen, D.M.: The relationship between variable selection and data augmentation and a method for prediction. Technometrics 16, 125–127 (1974)
Rivas, F.: Lignin and carbon transformation in roots of maize and mixed perennial biofuel crops. Graduate Theses and Dissertations Paper 12447, Iowa State University, Iowa, http://lib.dr.iastate.edu/cgi/viewcontent.cgi?article=3454&context=etd (2012). Accessed 15 April 2015
Lima, R.B., Salvador, V.H., dos Santos, W.D., Bubna, G.A., Finger-Teixeira, A., Soares, A.R., Marchiosi, R., Ferrarese, M.L., Ferrarese-Filho, O.: Enhanced lignin monomer production caused by cinnamic acid and its hydroxylated derivatives inhibits soybean root growth. PLoS ONE (2013). doi:10.1371/journal.pone.0080542
Kuo, B., Boersma, L.: Soil water suction and root temperature effects on nitrogen fixation. Agron. J. 63, 901–904 (1971)
Okmen, G., Donmez, G., Donmez, S.: Influence of osmotic and metal stresses on nitrogenase activity of cyanobacteria isolated from paddy fields. Afr. J. Biotechnol. 6, 1828–1832 (2007)
Dutta, N., Mukhopadhyay, A., Dasgupta, A.K., Chakrabarti, K.: Nanotechnology enabled enhancement of enzyme activity and thermostability: study on impaired pectatelyase from attenuated Macrophomina phaseolina in presence of hydroxyapatite nanoparticle. PLoS ONE (2013). doi:10.1371/journal.pone.0063567
Mukhopadhyay, A., Dasgupta, A.K., Chattopadhyay, D.J., Chakrabarti, K.: Improvement of thermostability and activity of pectatelyase in the presence of hydroxyapatite nanoparticles. Bioresour. Technol. (2012). doi:10.1016/j.biortech.2012.03.094
Konwarh, R., Karak, N., Rai, S.K., Mukherjee, A.K.: Polymer-assisted iron oxide magnetic nanoparticle immobilized keratinase. Nanotechnology (2009). doi:10.1088/0957-4484/20/22/225107
Navarro, E., Baun, A., Behra, R., Hartmann, N.B., Filser, J., Miao, A.J., Quigg, A., Santschi, P.H., Sigg, L.: Environmental behavior and ecotoxicity of engineered nanoparticles to algae, plants, and fungi. Ecotoxicology 17, 372–386 (2008)
Sun, H., Zhang, X., Niu, Q., Chen, Y., Crittenden, J.C.: Enhanced accumulation of arsenate in carp in the presence of titanium dioxide nanoparticles. Water Air Soil Pollut. 178, 245–254 (2007)
Poynton, H.C., Lazorchak, J.M., Impellitteri, C.A., Smith, M.E., Rogers, K., Patra, M., Hammer, K., Allen, J.H., Vulpe, C.D.: Differential gene expression in Daphnia magna suggests distinct modes of action and bioavailability for ZnO nanoparticles and Zn ions. Environ. Sci. Technol. (2011). doi:10.1021/es102501z
Bilberg, K., Hovgaard, M.B., Besenbacher, F., Baatrup, E.: In vivo toxicity of silver nanoparticlesand silver ions in zebrafish (Danio rerio). J. Toxicol. (2012). doi:10.1155/2012/293784
Misra, S.K., Dybowska, A., Berhanu, D., Luoma, S.N., Valsami-Jones, E.: The complexity of nanoparticle dissolution and its importance in nano-toxicological studies. Sci. Total Environ. 438, 225–232 (2012)
Neal, A.L., Kabengi, N., Grider, A., Bretsch, P.M.: Can the soil bacterium Cupriavidus necator sense ZnO nanomaterials and aqueous Zn2+ differentially? Nanotoxicol. 6(Suppl 4), 371–380 (2012)
Schwirn, K., Tietjen, L., Beer, I.: Why are nano-materials different and how can they be appropriately regulated under reach? Environ. Sci. Eur. (2014). doi:10.1186/2190-4715-26-4
Thill, A., Zeyons, O., Spalla, O., Chauvat, F., Rose, J., Auffan, M., Flank, A.M.: Cytotoxicity of CeO2 nanoparticles for Escherichia coli: physico-chemical insight of the cytotoxicity mechanism. Environ. Sci. Technol. 40, 6151–6156 (2006)
Tayel, A.A., El-Tras, W.F., Moussa, S., El-Baz, A.F., Mahrous, H., Salem, M.F., Brimer, L.: Antibacterial action of zinc oxide nanoparticles against food-borne pathogens. J. Food Safety (2011). doi:10.1111/j.1745-4565.2010.00287.x
Pramanik, A., Laha, D., Bhattacharya, D., Pramanik, P., Karmakar, P.: A novel study of antibacterial activity of copper iodide nanoparticle mediated by DNA and membrane damage. Colloids Surf. B (2012). doi:10.1016/j.colsurfb.2012.03.021
Cabiscol, E., Tamarit, J., Ros, J.: Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8 (2010)
Burello, E., Worth, A.P.: A theoretical framework for predicting the oxidative stress potential of oxide nanoparticles. Nanotoxicol. 5, 228–235 (2010)
George, S., Xia, T., Rallo, R., Zhao, Y., Ji, Z., Lin, S., Wang, X., Zhang, H., France, B., Schoenfeld, D., Damoiseaux, R., Liu, R., Lin, S., Bradley, K.A., Cohen, Y., Nel, A.E.: Use of a high-throughput screening approach coupled with in vivo zebrafish embryo screening to develop hazard ranking for engineered nano-materials. ACS Nano (2011). doi:10.1021/nn102734s
Seil, J.T., Webster, T.J.: Antimicrobial applications of nanotechnology: methods and literature. Int. J. Nanomed. 7, 2767–2781 (2012)
Zhang, H., Ji, Z., Xia, T., Meng, H., Low-Kam, C., Liu, R., Pokhrel, S., Lin, S., Wang, X., Liao, Y.P., Wang, M., Li, L., Rallo, R., Damoiseaux, R., Telesca, D., Mädler, L., Cohen, Y., Zink, J.I., Nel, A.: Use of metal oxide nanoparticle band cap to develop a predictive paradigm for oxidative stress and acute pulmonary inflammation. ACS Nano (2012). doi:10.1021/nn3010087
Sondi, I., Salopek-Sondi, B.: Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for gram-negative bacteria. J. Colloid Interface Sci. 275, 177–182 (2004)
Lynch, I., Dawson, K.A.: Protein–nanoparticle interactions. Nano Today 3, 40–47 (2008)
Saptarshi, S.R., Dusch, A., Lopata, A.L.: Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle. J. Nano-biotechnol. http://www.J.Nanobiotechnology.com/content/11/1/26 (2013). Accessed 15 April 2015
Mahmoudi, M., Lynch, I., Ejtehadi, M.R., Monopoli, M.P., Bombelli, F.B., Laurent, S.: Protein- nanoparticle interactions: opportunities and challenges. Chem. Rev. (2011). doi:10.1021/cr100440g
Chakraborti, S., Chatterjee, T., Joshi, P., Poddar, A., Bhattacharyya, B., Singh, S.P., Gupta, V., Chakrabarti, P.: Structure and activity of lysozyme on binding to ZnO nanoparticles. Langmuir 26, 3506–3513 (2009)
Sinha, R., Khare, S.K.: Differential interactions of halophilic and non-halophilic proteases with nanoparticles. Sustain. Chem. Process. (2014). doi:10.1186/2043-7129-2-4
Kathiravan, A., Paramaguru, G., Renganathan, R.: Study on the binding of colloidal zinc oxide nanoparticles with bovine serum albumin. J. Mol. Struct. 934, 129–137 (2009)
Miletic, N., Abetz, V., Ebert, K., Loos, K.: Immobilization of Candida antarctica lipase B on polystyrene nanoparticles. Macromol. Rapid Commun. 31, 71–74 (2010)
Colvin, V.L., Kulinowski, K.M.: Nanoparticles as catalysts for protein fibrillation. Proc. Natl. Acad. Sci. 104, 8679–8680 (2007)
Linse, S., Cabaleiro-Lago, C., Xue, W.-F., Lynch, I., Lindman, S., Thulin, E., Radford, S.E., Dawson, K.A.: Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 104, 8691–8696 (2007)
Nel, A., Madler, L., Velegol, D., Xia, T., Hoek, E., Somasundaran, P., Klaessig, F., Castranova, V., Thompson, M.: Understanding bio-physicochemical interactions at the nano-bio interface. Nat. Mater. 8, 543–557 (2009)
Wagner, S.C., Roskamp, M., Pallerla, M., Araghi, R.R., Schlecht, S., Koksch, B.: Nanoparticle induced folding and fibril formation of coiled-coil-based model peptides. Small 6, 1321–1328 (2010)
Deng, Z.J., Liang, M., Monteiro, M., Toth, I., Minchin, R.F.: Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat. Nanotechnol. 6, 39–44 (2011)
Acknowledgments
The authors are grateful to the Indian Council of Agricultural Research for funding the National Agricultural Innovation Project on nanotechnology (Project Code-C2032) under which the present work was conducted. The authors are also grateful to Shri Rajesh Kumar, Shri Mota Ram, Shri Praveen Kumar Chundawat and Shri Anil Kumar for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kumar, P., Burman, U. & Santra, P. Effect of nano-zinc oxide on nitrogenase activity in legumes: an interplay of concentration and exposure time. Int Nano Lett 5, 191–198 (2015). https://doi.org/10.1007/s40089-015-0155-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40089-015-0155-6