Abstract
Deletion of 13q14.3 and a candidate gene KCNRG (potassium channel regulating gene) is the most frequent chromosomal abnormality in B-cell chronic lymphocytic leukemia and is a common finding in multiple myeloma (MM). KCNRG protein may interfere with the normal assembly of the K+ channel proteins causing the suppression of Kv currents. We aimed to examine possible role of KCNRG haploinsufficiency in chronic lymphocytic leukemia (CLL) and MM cells. We performed detailed genomic analysis of the KCNRG locus; studied effects of the stable overexpression of KCNRG isoforms in RPMI-8226, HL-60, and LnCaP cells; and evaluated relative expression of its transcripts in various human lymphomas. Three MM cell lines and 35 CLL PBL samples were screened for KCNRG mutations. KCNRG exerts growth suppressive and pro-apoptotic effects in HL-60, LnCaP, and RPMI-8226 cells. Direct sequencing of KCNRG exons revealed point mutation delT in RPMI-8226 cell line. Levels of major isoform of KCNRG mRNA are lower in DLBL lymphomas compared to normal PBL samples, while levels of its minor mRNA are decreased across the broad range of the lymphoma types. The haploinsufficiency of KCNRG might be relevant to the progression of CLL and MM at least in a subset of patients.
Similar content being viewed by others
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) represents 22–30% of all leukemia cases, thus being the most commonly diagnosed form of leukemia in the Western world [1, 2]. This disease is usually seen amongst the elderly [2]. On a cellular level, the disease progresses due to the prolonged survival of B-CLL cells arrested in the G0 stage of the cell cycle [3, 4] This prolonged survival causes the accumulation of quiescent small CD5(+) B lymphocytes which are frozen at an early step of maturation and have decreased susceptibility to apoptotic cell death. Latter features of CLL are commonly attributed to the imbalance in various cytoplasmic pro-survival and pro-death pathways, including the BCR-signaling pathway [3, 5, 6]. In addition, a small number of “atypical” Ki-67 expressing lymphocytes are frequently observed in the bone marrow of CLL patients [7]. Proliferation rates of these lymphocytes vary among patients and even among subpopulations of cells within the leukemic clones of individual patients [8]. A newly evolving point of view at CLL stresses that blood lymphocyte count in individual patients represents a dynamic interplay between ongoing birth and death within the clones rather than a linear, monotonous accumulation of inert leukemic cells [8].
The most frequent chromosomal abnormalities in B-CLL are deletions on 13q14 and 17p13, trisomy 12, and rearrangements of 14q32. Being the most common abnormality in CLL [9, 10], deletions at 13q14.3 are associated with the longest survival. Rearrangements and/or deletions in the region of 13q14.3 are found in many other types of hematopoietic malignancies, including 38% of mantle cell lymphomas [11] and approximately 54% of multiple myelomas (MM) as detected by fluorescence in situ hybridization [12, 13]. In the majority of these non-CLL cases, 13q14 deletions are associated with a poor chemotherapy response profile. In addition, deletions of 13q14.3 are common in prostate cancer [14] and in some other solid tumors.
Attempts to delineate the minimal common deleted region (CDR) in CLL and MM cells lead to highlighting a region adjacent to marker D13S319 [9, 13, 15, 16]. This region encompasses an area containing the previously described tumor suppressor gene candidates DLEU1, DLEU2, RFP2, and KCNRG as well as microRNAs miR-15a and miR-16-1 [9, 16–19]. DLEU1 and DLEU2 candidates are represented by a number of non-coding RNAs [15, 20]. Direct sequencing and single-strand conformation polymorphism (SSCP) screening of the RFP2 gene encoding E3 ubiquitin ligase revealed no mutations [13, 19, 21]. Recently, a haploinsufficiency mechanism has been proposed as a major mode of inactivation of 13q14 tumor suppressor gene(s). According to this mechanism, deletion of a single active copy of 13q14.3 results in significant downregulation of the candidate gene(s) and loss of function [22].
Candidate tumor suppressor gene KCNRG (potassium channel regulating gene) is located within the 3′ end of the largest transcript of RFP2 [23] and encodes a protein with a high homology to the tetramerization domain of voltage-gated K+ channels (Kv channels). This protein may interfere with the normal assembly of the K+ channel proteins by binding to their tetramerization domain, thereby, causing the suppression of Kv currents. This hypothesis was consequently confirmed using the patch-clamp technique in the human androgen-sensitive prostate cell line LnCaP [23]. As Kv channels are involved in the proliferation of both LnCaP cells [24] and normal lymphocytes [25], while being upregulated in neoplastic hematopoietic cells [26], we hypothesized that KCNRG may exert a tumor suppressor effect relevant to CLL and MM.
To investigate the possibility of KCNRG involvement in the pathogenesis of these diseases, we performed detailed genomic analysis of the KCNRG locus; studied effects of the stable overexpression of KCNRG isoforms in the model cell lines RPMI-8226, HL-60, and LnCaP; and evaluated relative expression of its transcripts in various human lymphomas.
Materials and methods
Mutation screening
This study was performed using peripheral blood samples obtained from 35 B-CLL patients and ten donors with informed consent and Institutional Review Board approval. Peripheral blood mononuclear cells were isolated by a standard procedure using Ficoll-Hypaque gradient centrifugation and used for total DNA preparation and purification by phenol/chlorophorm protocol [27]. Median absolute number of lymphocytes was 57,000/ul (range 24,000–187,000); therefore, in all cases, fraction of mononuclear cells contained mostly B-CLL cells. Primer sequences for amplifying the coding region of the KCNRG gene were chosen according to Cho et al. All cases were screened for the presence of an aberrant band in the tumor DNA by SSCP analysis of each exon and compared with the normal DNA. Highly polymorphic repeat (A)33(TA)8(CA)8TATGTA(CA)2TACA (TA)4CA(TA)3 has been amplified with primers 5′-GTA AAC ATA GCA AGA CCC AGT-3′ and 5′-CAT TTC CTG CTC TTG CCA TGT-3′; average amplicon length was 158 bp. KCNRG exons along with adjacent areas were directly amplified on the DNA templates of MM cell lines NCI-H929, RPMI-8226, and U266 and subjected to automated sequencing.
Cloning of human KCNRG isoforms
All basic molecular biology and microbiology procedures (plasmid isolation, cloning, Escherichia coli transformation, and cultivation) were performed according to standard protocols [27]. Human KCNRG-L (mRNA isoform B) and KCNRG-S (mRNA isoform A) were cloned into the pcDNA3.1/myc-His vector (Invitrogen, San Diego, CA, USA). The cloning of KCNRG-L was performed by OriGene (Rockville, MD, USA) using archived full-length cDNA clone corresponding to mRNA isoform NM_173605. For KCNRG-S isoform, a polymerase chain reaction (PCR) product encompassing its full-length open reading frame was amplified from human brain mRNA, cloned into pGemT-Easy vector (Promega), then transferred to pcDNA3.1/myc-His. The restriction enzymes BamH1 and XhoI were used for the cloning of both KCNRG isoforms. Bidirectional sequencing of the plasmids and PCR clones with vector or gene-specific primers were performed by fluorescence-tagged chain termination (Big Dye Terminator, Applied Biosystems), according to the manufacturer's protocol, followed by fractionation on an ABI 310 automated DNA sequencer (Applied Biosystems). Two primer pairs were designed to confirm authenticity of the inserts: pair V1 capable of amplifying both isoforms of KCNRG with product sizes of 107 basepairs and 129 basepairs, and pair V2, specifically amplifying KCNRG-S specific product of 129 basepairs. Primer sequences were as follows: V1 5′-TTTTCCCTCCTCAGATGACC-3′ and 5′-TCCAGTTTGGTTATCAGTAGTGC-3′, V2 5′-CCTGGTTTTCCAGTGTG GTT-3′, and 5′-GCTGAGGCAGGAGAATCACT-3′.
LnCaP, HL-60, and RPMI-8226 cells stably expressing KCNRG-L and KCNRG-S
Cell lines LNCaP, HL-60, and RPMI-8226 were obtained from American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI-1640 containing 2 mM glutamine, 10 mM HEPES, and 10% fetal calf serum (Invitrogen, San Diego, CA, USA). The cDNA-containing or empty control plasmids were transfected into LNCaP, HL-60, and RPMI-8226 cells with Transfectol (Gene Choice, Frederick, MD, USA). Stably transfected cells were selected using 500 ug/ml of Geneticin (Sigma) and were subsequently maintained in 75 cm2 flasks with antibiotic. Expression of KCNRG isoforms was confirmed by real-time PCR as described below.
Cell proliferation and apoptosis assays
For cell proliferation and apoptosis assays, cells were seeded in 96-well black plates with clear bottoms (Thermo Electron) at a density of 2 × 104 per well (proliferation assay) or 3.5 × 104 per well (apoptosis assay) in 100 ul of RPMI-1640 media with 10% fetal calf serum. For the proliferation assays, 32 wells were seeded with stably transfected cells, vector control cells, and wild-type parental cell line along with eight no-cell background controls. In experiments with transiently transfected cells, an extra set of 16 wells with mock-transfected cells were added. Apoptosis assays were performed in 96 well plates in sets of 8-well replicates for each cell type. Cell proliferation rates were quantified by enzyme-linked immunosorbent assay (ELISA) Chemiluminescent BrDU (Roche). As recommended by the manufacturer, the optimal incubation times were determined experimentally, 48 h for RPMI-8662 and HL-60 and 72 h for LnCaP. Apoptosis rates were determined using the CaspaseGLO 3/7 luminescent Assay (Promega, Madison, WI, USA) that measures caspase-3 and -7 activities after 2 h of incubation. Chemoluminescence was quantified using microplate fluorometer Fluoroskan Ascent (Thermo Scientific). Both apoptosis and cell proliferation assays were performed in eight replicates.
FACS analysis
Cell cycle distribution was analyzed by flow cytometry as described earlier [28]. Briefly, 0.5 × 106 to 1.5 × 106 cells were transfered to 15 ml conical tubes, washed twice with cold phosphate buffer solution (PBS), centrifuged at 1,000 g for 5 min, and resuspended in 1 mL of PI staining solution (PBS + 50 ug/ml PI + 100 ug/ml RNAse A). Cells were incubated at 4°C in the dark for 20 min and were analyze on FACSCalibur (Becton Dickinson) within 15–30 min. Quantification of cell death was performed by staining with Annexin V/7-AAD kit (BD Pharmingen, San Jose, CA, USA). Washed and centrifuged pellets were resuspended in 0.1 ml of 1× binding buffer, then 10 µl of annexin V and 5 µl of 7-AAD were added to each sample. Cells were gently pipetted and incubated at room temperature for 15 min in the dark. Following incubation, 0.9 ml of 1× binding buffer was added to the cells. The samples were analyzed within 1 h of staining. Controls stained with Annexin V only and 7-AAD only were performed for calibration purposes. During the experiments, cells were kept on ice and away from light sources.
Cell imaging
Cells were rinsed in PBS briefly to remove media components and fixed in freshly prepared 4% paraformaldehyde/PBS solution for 15 min at room temperature. Cells were permeabilized by adding 0.1% Triton-X100 in PBS for 1 min. A PBS-diluted solution of Rhodamine phalloidin (1:100 in PBS), 5 uL of 300 nM DAPI, and 4 uL of diluted DIOC 18 solution were added to the permeabilized cells. The mixture was incubated for 15 min at room temperature. Following staining, the cells were rinsed in PBS three times and mounted onto slides with cover slips; slides were kept in the dark and in the freezer until imaging. Images were taken on a Nikon Eclipse 90i microscope equipped with a Nikon C1 confocal scan head and laser lines at 406, 488, and 568 nm.
Invasion and migration assays
To evaluate migratory and invasive properties of the cells overexpressing KCNRG, fluorimetric CytoSelect 96well Cell Migration and Invasion, CytoSelect 24-wells Anoikis, and CytoSelect Leukocyte Transmigration Assays (Cell Biolabs, Inc, San Diego, CA, USA) were used according to the manufacturer's protocols. Invasion and migration assays were performed in three replicates.
Proteome analysis
For reverse-phase protein microarrays methodology, the protein lysates were loaded into 384-well plates and each serially diluted in lysis buffer to a five-point dilution curve (neat, 1/2, 1/4, 1/8, and 1/16). Each dilution series was printed in duplicate onto nitrocellulose-coated glass slides (Whatman, Inc., Sanford, ME, USA) with a 2470 Arrayer (Aushon BioSystems, Burlington, MA, USA). Slides were desiccated and stored at −20°C. Before antibody staining, the lysate arrays were treated with mild Reblot antibody stripping solution (Chemicon, Temecula, CA, USA) for 15 min at room temperature, washed twice for 5 min in phosphate-buffered saline, and then incubated for at least 5 h in blocking solution [1 g I-block (Tropix, Bedford, MA, USA), 0.1% Tween-20 in 500 ml phosphate-buffered saline] at room temperature with constant rocking. Blocked arrays were stained with antibodies on an automated slide stainer (Dako Cytomation, Carpinteria, CA, USA) using the Catalyzed Signal Amplification System kit according to the manufacturer's recommendation (CSA; Dako Cytomation). Briefly, endogenous biotin was blocked for 10 min with the biotin blocking kit (Dako Cytomation), followed by application of protein block for 5 min; primary antibodies were diluted in antibody diluent and incubated on slides for 30 min, and biotinylated secondary antibodies were incubated for 15 min. Signal amplification involved incubation with a streptavidin–biotin–peroxidase complex provided in the CSA kit for 15 min and amplification reagent (biotinyl-tyramide/hydrogen peroxide, streptavidin-peroxidase) for 15 min each. Development was completed by using diaminobenzadine/hydrogen peroxide as the chromogen/substrate. Slides were allowed to air dry after development. We specifically chose 38 primary antibodies to analyze broad signaling pathways thought to be involved in the proliferation and apoptosis-related signaling. Secondary antibodies and dilutions included biotinylated goat anti-rabbit IgG (H_L) 1:5,000 (Vector Laboratories, Burlingame, CA, USA) and biotinylated rabbit anti-mouse IgG 1:10 (Dako Cytomation). Stained slides were scanned individually on a UMAX PowerLook III scanner (UMAX, Dallas, TX, USA) at 600 dpi and saved as TIF files in Photoshop 6.0 (Adobe, San Jose, CA, USA). The TIF images for antibody stained slides and Sypro-stained slide images were analyzed with MicroVigene image analysis software, version 2.200 (Vigenetech, North Billerica, MA, USA), and Microsoft Excel 2000 software. Images were imported into Microvigene, which performed spot finding, local background subtraction, replicate averaging, and total protein normalization, producing a single value for each sample at each endpoint (see Supplementary Table 1 for antibody details).
Real-time PCR profiling of KCNRG in human lymphoma samples and cell lines
To assess KCNRG gene expression in human lymphoma samples, TissueScan Tissue qPCR Arrays Panels were purchased from OriGene Technologies, Inc (Rockville, MD, USA). The pre-normalized cDNAs per each panel were composed of six normal, ten follicular lymphomas, 11 diffuse large B-cell lymphomas, three small lymphocytic lymphomas, six Hodgkin lymphomas, eight marginal zone B-cell lymphomas, two mantle cell lymphomas, and three T-cell lymphomas. Reactions were performed in a 96-well format in 30 uL reactions in the BioRad iCycler iQ Real-Time Detection System (BioRad Laboratories, Hercules, CA, USA). For KCNRG isoforms, three independent PCR experiments on individual pre-normalized Tissue Scan plates from the same manufacturer's batch were performed. Primers used for real-time PCR profiling of KCNRG isoforms were KCNRG-SL primers 5′-TTTTCCCTCCTCAGATGACC-3′ and 5′-TCCAGTTTGGTTATCAGTAGTGC-3′; KCNRG-S primers 5′-CCTGGTTTTCCAGTGTGGTT-3′ and 5′-GCTGAGGCAGGAGAATCACT-3′. Additional normalization was performed in triplicate using Beta Actin control primers provided by the manufacturer. The presence of a single specific PCR product was verified by melting curve analysis and confirmed on agarose gels.
For an assessment of endogenous and overexpressed levels of KCNRG transcripts, total RNA preparates were isolated by Qiagen RNeasy Mini Kit (Valencia, CA, USA) from wild-type cell lines LNCaP, HL-60, and RPMI-8226 (ATCC, Manassas, VA, USA) as well as from their derivatives stably expressing KCNRG-S, KCNRG-L, and pcDNA3.1 backbone control. Reverse transcription reactions were performed using 2 μg of total RNA and SuperScript® Reverse Transcriptase (RT; Invitrogen, San Diego, CA, USA). Reactions were heated at 70°C for 5 min in a total volume of 12.5 μl in the presence of 100 ng of random hexamers (Invitrogen) and cooled at room temperature for 1 min. After a mini-centrifuge spin, 4 μl of 5× first strand buffer (Invitrogen), 2 μl of 0.1 M DTT, and 0.5 μl of 25 mM dNTP mix (Fisher Scientific) were added to the reaction. The mixture was then incubated at 42°C for 1 h. The resulting cDNA was stored frozen (−80°C) until assayed by real-time PCR. PCR reactions were performed in a 96-well format in the BioRad iCycler iQ real-time Detection System (BioRad Laboratories, Hercules, CA, USA). The real-time PCR mixtures contained 1 μl of the RT sample, 400 nM each of forward and reverse primers, and 1× iQ SYBR Green Super Mix and were carried out in a total volume of 15 μl. Amplification of 18S RNA in parallel with the genes of interest was performed as an internal normalization standard with previously described primers 18S-F 5′-AGGAATTCCCAGTAAGTGCG-3′ and 18S-R 5′-GCCTCACTAAACCATCCAA-3′ [19, 29]. For each gene of interest and 18S RNA, three to four independent PCR experiments from the same RT sample were performed.
Computer programs and statistical analysis
Comparative analysis in silico and an analysis of the structure of the KCNRG gene were performed using genomic, mRNA and EST databases (http://www.ncbi.nlm.nih.gov and http://genome.ucsc.edu). The KCNRG promoter was predicted by Core Promoter (http://www.cshl.edu/OTT/html/corepromoter.html) and NNPP (http://www.fruitfly.org/seq_tools/ promoter.html) services. Search for known single nucleotide polymorphisms (SNPs) was performed in National Center for Biotechnology Information dbSNP build 129. A search for TF binding sites and promoter elements was performed using MatInspector algorithm and Matrix Family Library Version 7.1 (June 2008 release, http://www.genomatix.de/). MAR/SAR elements were predicted by SMARTest (www.genomatix.de). Multiple alignments of the protein sequences were performed using ClustalW v.1.83. A phylogenetic tree of prealigned T1 domains of KCNRG-like proteins was reconstructed by the neighbor-joining method and visualized by the same software. Group comparisons were performed by non-parametric Mann–Whitney hypothesis tests. Unless otherwise noted, P values <0.05 were considered significant.
Supplementary materials
Supplementary Figures 1 and 2 as well as Supplementary Table 1 could be downloaded at http://mason.gmu.edu/∼abaranov/pages/completed%20project%20pages/Compl-KCNRG.html.
Results
mRNA and protein isoforms of KCNRG gene
Candidate tumor suppressor gene KCNRG resides within the 3′ end of the largest transcript of RFP2 gene described earlier [19, 30]. Major mRNAs of KCNRG are transcribed independently of RFP2, starting at the promoter located within 3′-untranslated region RFP2 (Fig. 1). This sequence is adjacent to in silico predicted promoter located in the position approximately 100 nt upstream of the putative 5′ end of the KCNRG transcripts according to an alignment of the KCNRG ESTs to genome (Core Promoter score 1.000, NNPP score 0.97). Additionally, RT-PCR experiments support existence of a hybrid mRNA isoform that includes exons from both RFP2 and KCNRG (Fig. 1). This isoform originates from the quadruplex containing promoter of RFP2, possibly due to its unusual properties [31]. In all examined species of mammals with the exception of primates, KCNRG and RFP2 genes are encoded by separate loci (Supplementary Figure 1). Prediction of MAR/SAR elements that exhibit enhanced affinities for nuclear matrix binding does not reveal any of these in mouse locus and only one such element in the intron of RFP2 in rat genome, while KCNRG/RFP2 locus in human genome contains five of these elements, possibly indicating substantial differences in the principles of the regulation of these genes in humans and rodents.
Genomic organization of RFP2/KCNRG gene locus. Schemes represent the structure of the mRNA isoforms of the human RFP2 and KCNRG genes and the hybrid mRNA isoform. Open reading frame of RFP2 is represented by white arrow. Open reading frames of KCNRG are represented by black arrows. Hybrid mRNA RFP2/KCNRG is not translated. Promoter of RFP2 marked as PR, promoter of KCNRG marked as PK
Human KCNRG encodes two protein isoforms KCNRG-L (272 aa) and KCNRG-S (229aa) differing in their C-ends and possessing common N-end of 184 aa. A T1 tetramerization domain covers amino acid positions 7 to 98. KCNRG loci of non-human mammals encode only one protein isoform corresponding to human KCNRG-L. In chimps, KCNRG-L differs from its human orthologue by one amino acid substitution (Pro → Leu) in the position 158. Comparison of human and rat KCNRG orthologues revealed 85.4% identity in 268 residue overlap, while comparison with mouse orthologue was characterized by 73.2% identity in 264 residue overlap. Murine KCNRG locus encodes two protein isoforms, 264 and 191 residues in length, both of which are variants of human KCNRG-L isoform.
Interestingly, human KCNRG-S and KCNRG-L isoforms are different by their C-tails, as these proteins share only first 191 amino acids. N-end difference is due to out-of-frame insertion of the alternatively spliced exon 2 that is present only in the human genome and is derived from AluSp SINE repeat. Human mRNA isoforms encoding two KCNRG proteins are co-expressed in the same set of tissues (not shown). Levels of Alu-containing KCNRG-S mRNA isoform are substantially lower than that of KCNRG-L mRNA.
KCNRG is a member of the KCTD protein family
Human KCNRG is a member of the KCTD protein family that encodes predicted proteins with an N-terminal domain homologous to the T1 domain in voltage-gated potassium channels. KCTD family proteins belong to a larger group of non-channel T1/BTB proteins. KCTD family members are similar to Pfam K_tetra consensus (PF02214) rather than that of BTB/POZ [32]. Our analysis (Fig. 2; Supplementary Figure 2) revealed two previously unclassified KCNRG-like proteins that belongs to KCTD family, TNFAIP1 (tumor necrosis factor-alpha-induced protein 1) and SHKPB1 (SH3KBP1 binding protein 1), while pointing that the protein C6orf69 previously described as KCTD20 represents an outgroup and should be excluded from the T1 family in a sensu stricto. Most of the KCTD proteins are relatively short (on average, 355 aminoacids). Their T1 domains are located close to the N-end of the protein and are not accompanied by any other Pfam domains. KCTD19 protein is the only KCTD member that contains two tandem T1 domains, first of which is truncated. T1 domains in three out of 25 KCTD proteins, namely, KCTD18, KCTD11, and KCTD9L, are truncated. KCNRG is a typical member of the KCTD family, with solitary T1 domain unaccompanied by any other recognizable protein structure.
An alignment of KCNRG with other proteins of KCTD family. Degree of shading indicates different degree of conservation for a given amino acid position: invariant positions are darkest, other conserved positions are shaded lighter, and non-conserved positions are not shaded. Total length of T1 domain, its position, and full length of the proteins are summarized in the table adjacent to the aligned protein sequences. Truncated versions of T1 domain are marked by star
Variations in KCNRG locus
Mutations within open reading frame of KCNRG and adjacent parts of its introns were screened in 35 CLL patients by conventional PCR-SSCP. No aberrant bands were seen. Direct sequencing of KCNRG exons revealed point mutation delT in the +30 position of the 5′ non-coding area of the gene in RPMI-8226 cell line (position indicated according to mRNA Ac. Num. NM_199464). The search for known human SNPs was negative for any match in this nucleotide position. Interestingly, an analysis of matches to transcription factor binding matrices with MatInspector software revealed the core promoter initiator element (INR) that overlaps with this mutated position. Deletion of T decreases matrix similarity of the match from 0.945 to 0.941, and therefore, might negatively influence expression of KCNRG in RPMI-8226 cells.
A low complexity repeat (A)33(TA)8(CA)8TATGTA(CA)2TACA(TA)4CA(TA)3 located at the position 1006 relative to the major start of KCNRG mRNAs and within 3′ untranslated area of RFP2 was amplified by PCR with subsequent band resolution in polyacrilamide gel in 21 DNA sample of CLL patients and 50 DNA samples of normal donors. Unusually high level of the polymorphism has been observed, as the profiling of a total of 142 chromosomes revealed 12 alleles of this repeat. Heterozygous state of this repeat was seen in 52/71 (73%) samples tested indicating high informativeness of this polymorphic DNA marker.
Expression of KCNRG inhibits proliferation and enhances apoptosis in cultured tumor cells
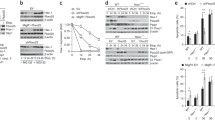
Open reading frames corresponding to both protein isoforms of KCNRG were stably transfected in LNCaP, HL-60, and RPMI-8226 cells and studied in various functional assays. The cells transfected with pcDNA3.1 vector alone were used as a control. As quantified by chemiluminescent BrdU cell proliferation ELISA assay, growth of all three cell lines was significantly inhibited by an overexpression of KCNRG-L isoform (RPMI-8226: decrease of 37%, P < 0.001; HL-60: decrease of 26%, P < 0.0025; LNCaP: decrease of 38%, P < 0.009). KCNRG-S isoform exerted less prominent growth suppressive effect in these cells (RPMI-8226: decrease of 27%; HL-60: decrease of 12%; LNCaP: decrease of 24%). In the case of KCNRG-S isoform, the differences between observed values were significant only for KCNRG-S overexpression in LNCaP (P < 0.05) and RPMI-8226 (P < 0.02; Fig. 3a–c).
Expression of KCNRG inhibits proliferation and enhances apoptosis. Axis Y reflects chemoluminescence readings (ChemU) in respective assays. a–c Results BrdU enzyme-linked immunosorbent assay to quantify proliferation in HL-60 (a), RPMI-8226 (b), and LnCaP (c) cells. d–f Results of the caspase assays to quantify apoptosis in HL-60 (d), RPMI-8226 (e), and LnCaP (f) cells
Cell survival was analyzed by measurement of the activities of caspases 3 and 7 (Fig. 3d–f). We revealed an increase in the apoptotic events in the cells that were stably transfected with any KCNRG isoform, but in KCNRG-L cells, these changes were more profound (KCNRG-L/RPMI-8226: increase of 180%, P < 0.0008; KCNRG-L/HL-60: increase of 216%, P < 0.0008; KCNRG-L/LNCaP: increase of 46%, P < 0.0002; KCNRG-S/RPMI-8226: increase of 94%, P < 0.0008; KCNRG-S/HL-60: increase of 25%, NS; KCNRG-S/LNCaP: increase of 41%, P < 0.0002).
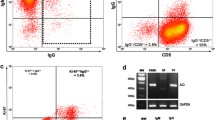
Observations described above were confirmed by FACS analysis of the cell lines overexpressing KCNRG isoforms and the vector control (Figs. 4 and 6). Interestingly, the suppression of the proliferation of the suspension cell lines HL-60 and RPMI-8226 has been accompanied by the dramatic changes in the size and the shape of the cells. An increase of KCNRG-L expression in HL-60 cell line resulted in the formation of two cell populations, normal-like population mainly residing in G1 phase (Fig. 4f, g, quadrants 1 and 3) and larger, abnormal cells residing in G2 (Fig. 6f, g, quadrants 2 and 4). Similarly, an overexpression of KCNRG-L in RPMI-8226 also produced a population of abnormal cells (not shown). Cell imaging studies indicated a change of morphology in KCNRG overexpressing cells and generally confirmed FACS findings (Figs. 5 and 6).
Propidium iodide FACS analysis of the cell cycle in the vector control and KCNRG-L overexpressing HL-60 cells. a, b Scatter plot and frequency histogram reflecting cell sizes and DNA content in HL-60 cells transfected with empty vector (vector control). c, d Scatter plot and frequency histogram reflecting cell sizes and DNA content in HL-60 cells transfected with KCNRG-L overexpressing plasmid. e Overlay of images (a) and (c). f, g Detailed frequency histograms displaying DNA content of vector control HL-60 cells (f) and KCNRG-L overexpressing HL-60 cells (g) located by their size in each of four quadrants
Images of HL-60 cells. a–d HL-60/control. e–h HL-60/KCNRG-L. i–l RPMI-8226/control. m–p RPMI-8226/KCNRG-L. Cells stained with lipohilic probe Dioc 18 (a, e, i, m), Nucleic acid probe DAPI (b, f, j, n), and F-actin probe rhodamine phalloidin (c, g, k, o). Overlay of the individual images (d, h, l, p). Original magnification ×100
Quantification of cell death in HL-60/control, HL-60/KCNRG-L, and HL-60/KCNRG-S by staining with Annexin V/7-AAD. a–c Scatter plots and frequency histograms reflecting cell sizes and staining with Annexin V/7-AAD in HL-60 cells vector control (a), HL-60 cells transfected with KCNRG-L overexpressing plasmid (b), and HL-60 cells transfected with KCNRG-L overexpressing plasmid (c). d Overlay of images (a) and (b). e Overlay of images (a) and (c). f Bar graph reflecting relative changes in the number of cells stained with Annexin V/7-AAD after transfection with KCNRG isoforms. Q1 reflects necrosis, Q2 reflects late apoptosis, Q3 reflects healthy cell population not affected by apoptosis or necrosis, and Q4 reflects early apoptosis
Expression of KCNRG does not influence invasion of LNCaP cells but affects migration of the suspension cell lines HL-60 and RPMI-8226
Migration of RPMI-8226 cells stably transfected KCNRG-S and KCNRG-L as well as vector control has been studied using 0.2–2.0 × 106 cells/ml and fetal bovine serum as chemoattractant. Migration of KCNRG-L overexpressing cells were not different from that of the control cells, while migration abilities of KCNRG-S were significantly lower (KCNRG-S, N = 28, 3.46 ± 0.408 vs. control, N = 28, 2.75 ± 0.67, P < 0.004). Analysis of the transmigration of the vector control and KCNRG overexpressing HL-60 cells through the monolayer of endothelial cell revealed that KCNRG-L decreases ability of the cells to penetrate endothelium (KCNRG-L, N = 5, 17.19 ± 0.26 vs. control, N = 5, 18.41 ± 0.59, P < 0.01), while overexpression of KCNRG-S leads to a non-significant increase in the transmigratory abilities of this cell line. Study of the adherent LNCaP cells revealed that overexpression of the KCNRG isoforms does not influence their ability to cross basement membrane (not shown). Study of LNCaP anoikis showed that both KCNRG overexpressing and vector control cells are dying upon detachment, thus indicating no influence of KCNRG on the anoikis related cell death (not shown).
Proteome changes associated with KCNRG overexpression
To evaluate differences in intracellular molecular networks associated with overexpression of KCNRG, we attempted RPA profiling of the changes in the broad spectrum of the cellular signaling events, with emphasis on the phosphorylation and proteolytic events associated with proliferation and apoptosis. In all three cell lines, an overexpression of either isoform of KCNRG was associated with decreased activation of mTOR by phosphorylation of serines 2481 and 2448 and decreased phosphorylation of tyrosine 1175 in VEGFR2. In RPMI-8226, overexpression of KCNRG isoforms lead to the coordinated increase of cleavage events in caspases 3, 6, 7, and 9 and the caspase target protein PARP. In LNCaP cells, overexpression of KCNRG resulted in paradoxical profound increase in ERK signaling as revealed by the phosphorylation of Threonine 202 and Tyrosine 204.
Real-time PCR profiling of KCNRG mRNAs in human lymphoma samples
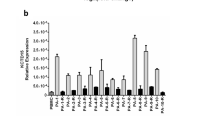
Relative levels of KCNRG isoforms were quantified in human lymphoma samples arrayed on TissueScan qPCR panels (Fig. 7). As expression of the major isoform of KCNRG mRNA encoding KCNRG-L was 300 times higher than that of the minor mRNA isoform (KCNRG-S), mRNA level estimates obtained using primer pair KCNRG-SL can be approximated to the expression of KCNRG-L encoding isoform. We revealed significant downregulation of KCNRG-L mRNA (P < 0.05) in the most advanced lymphomas of the stage IV (N = 4) as compared to normal PBL samples (N = 6). When the same set of samples were stratified according to the type of lymphoma, a significant downregulation of KCNRG-L was registered only in DLBL (N = 11) as compared to normal PBL samples (P < 0.002). Interestingly, only one case of DLBL was classified as stage IV disease, while six of these cases were stage I, and four were staged as IE or IIE. A trend towards downregulation of the KCNRG-S encoding mRNA was seen in all lymphoma types profiled, with statistically significant differences seen in FL (P < 0.002), DLBL (P < 0.02), and T-cell lymphoma (P < 0.03) groups. Stage-wise, significant decrease in the level of mRNA for KCNRG-S was seen in stage I (P < 0.008) and IIE (P < 0.04) groups.
Real-time polymerase chain reaction (PCR) profiling of KCNRG mRNAs in human lymphoma samples. N Normal PBL, FL follicular lymphoma, DLBL diffuse large B-cell lymphoma, SLL small lymphocytic lymphoma, HL Hodgkin lymphoma, MZBL marginal zone B-cell lymphoma, MCL mantle cell lymphoma, TL T-cell lymphoma. a, b Reverse transcription (RT)-PCR profiling using primer pair KCNRG-SL that amplifies both isoforms of KCNRG mRNA. c, d RT-PCR profiling using primer pair KCNRG-S that amplifies only mRNA isoform KCNRG-S. Statistically significant changes denoted by star symbol. Expression levels of KCNRG-L/S and KCNRG-S isoforms are reflected at Y axis in artificial expression units (AU) after normalization to β-actin mRNA levels
Discussion
Previous studies of the tumor suppressor effects conveyed by presence of an intact chromosome 13 region q14.3 revealed a number of potential candidate genes whose expression is lost in a substantial percentage of CLL cases [9, 16–19, 33]. Despite significant effort undertaken by multiple laboratories, a screening of the mutations in the studied candidate genes in primary tumor cells has not yet been successful. These findings prompted some investigators to propose that a major mode of inactivation of 13q14 tumor suppressor gene(s) is by the deletion of one copy of the gene causing decrease in the production of gene' mRNA and partial loss of its function [21, 22]. This mechanism gene inactivation is known as haploinsufficiency. Importantly, haploinsufficiency implies that the deleted region may contain more than one gene participating in the suppression of the tumor development, a feature that is especially relevant in light of particular importance of the conservative miRNA-16 locus for the development of CLL like disorder in New Zealand mice [33].
Candidate tumor suppressor gene KCNRG (potassium channel regulating gene) is located very close to the 10-kb region previously described as CDR in CLL and within larger region deleted in the majority of CLL cases with 13q14.3 aberrations [9, 23]. Functional analysis of KCNRG [23] pointed at the possibility that it might exert a tumor suppressor effect relevant to CLL and MM. Additionally, a missense mutation at codon 92 within the T1 domain of KCNRG was found in one hepatocellular tumor [34]. Analysis of the Hep3B hepatoma cells revealed that in this cell line, KCNRG has growth suppressive activity [34]. In this paper, we demonstrated that KCNRG possesses similar activity when overexpressed in RPMI-8226 and HL-60 cells. Moreover, overexpression of KCNRG stimulates apoptosis in these cells and leads to dramatic changes in their size and shape. A substantial proportion of both RPMI-8226 and HL-60 cells overexpressing isoform KCNRG-L was arrested in G2 phase, possibly indicative of G2/M checkpoint activation. Additionally, migration of KCNRG overexpressing cells has also significantly decreased. Finally, KCNRG overexpression increased a propensity of the cells to undergo apoptosis after the stress, in particular, affecting its ability to recover after the storage in frozen state (not shown).
One way to confirm the relevance of these characteristics of human KCNRG to the suppression of CLL and MM in vivo is to find its point mutations in the primary tumor cells. Our and others' attempts to do so were unsuccessful. However, a number of circumstantial evidences points at KCNRG as an important player in hematological malignancies. First, real-time PCR profiling of KCNRG mRNAs revealed that levels of the major KCNRG isoform in DLBL lymphomas are lower compared to normal PBL samples, while levels of its minor mRNA are decreased across the broad range of the lymphoma types. Levels of the KCNRG mRNA were also decreased in the lymphomas of the stage IV. Second, MM cell line RPMI-8226 contains delT mutation in the core promoter initiator element that might influence levels of KCNRG expression in this model line. Third, proteomics endpoint analysis by RPA indicated an involvement of KCNRG in the suppression of mTOR and VEGFR pathways as well as in caspase activation in a broad sense. In light of these observations, an identification of the particular mode of KCNRG inactivation in the tumor samples becomes a priority. Despite a number of studies that delineated 13q14 deletions in CLL and MM and quantified relative expression levels of some candidate genes residing in this area, the critical insights related to the role of 13q14 role in human malignancies are still to be gained. Here, we described a novel, highly informative polymorphic repeat, located at the position 1006 relative to the major start of KCNRG mRNAs and within 3′ untranslated area of RFP2. As the structure of this repeat allows its relatively simple visualization in agarose gels after conventional PCR, it might become a valuable marker for the hemizygous deletion of KCNRG in primary tumor samples.
Interestingly, KCNRG belongs to rather large subfamily of the poorly characterized human genes (Fig. 2). Some of these genes functions in tumorigenesis: SH3KBP1 suppresses cathepsin B-mediated apoptosis [35]; hedgehog antagonist REN (KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors and suppresses medulloblastoma genesis [36]; and expression of the KCTD12 (pfetin) serves as a negative prognostic biomarkers for gastrointestinal stromal tumors [37]. As cellular functions for the majority of the KCNRG-like proteins have never been studied, it is important to determine their relevance to the tumorigenesis and lymphomagenesis in humans.
In conclusion, we conducted a functional study of the tumor suppressor gene candidate KCNRG and demonstrated its growth suppressive and pro-apoptotic effects in the cellular models relevant to CLL and MM. We conclude that the effects of the loss of KCNRG might be relevant to the progression of these hematological malignancies at least in a subset of the patients with these disorders.
References
Caporaso N, Marti GE, Goldin L. Perspectives on familial chronic lymphocytic leukaemia: genes and the environment. Semin Hematol. 2004;41:201–6.
Redaelli A, Laskin BL, Stephens JM, Botteman MF, Pashos CL. The clinical and epidemiological burden of chronic lymphocytic leukaemia. Eur J Cancer Care (Engl). 2004;13:279–87.
Danilov AV, Danilova OV, Klein AK, Huber BT. Molecular pathogenesis of chronic lymphocytic leukaemia. Curr Mol Med. 2006;6:665–75.
Gale RP, Foon KA. Biology of chronic lymphocytic leukaemia. Semin Hematol. 1987;24:209–29.
Ougolkov AV, Bone ND, Fernandez-Zapico ME, Kay NE, Billadeau DD. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukaemia B cells. Blood. 2007;110:735–42.
Iglesias-Serret D, de Frias M, Santidrian AF, Coll-Mulet L, Cosialls AM, Barragán M, et al. Regulation of the proapoptotic BH3-only protein BIM by glucocorticoids, survival signals and proteasome in chronic lymphocytic leukaemia cells. Leukemia. 2007;21:281–7.
Bueso-Ramos CE, Ferrajoli A, Medeiros LJ, Keating MJ, Estrov Z. Aberrant morphology, proliferation, and apoptosis of B-cell chronic lymphocytic leukaemia cells. Hematology. 2004;9:279–86.
Chiorazzi N, Ferrarini M. Evolving view of the in-vivo kinetics of chronic lymphocytic leukaemia B cells. Hematology Am Soc Hematol Educ Program. 2006;273–8:512.
Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grandér D, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukaemia. Oncogene. 1997;15:2463–73.
Dohner H, Stilgenbauer S, Dohner K, Bentz M, Lichter P. Chromosome aberrations in B-cell chronic lymphocytic leukaemia: reassessment based on molecular cytogenetic analysis. J Mol Med. 1999;77:266–81.
Rosenwald A, Ott G, Krumdiek AK, Dreyling MH, Katzenberger T, Kalla J, et al. A biological role for deletions in chromosomal band 13q14 in mantle cell and peripheral t-cell lymphomas? Genes, Chromosomes Cancer. 1999;26:210–4.
Harrison CJ, Mazzullo H, Cheung KL, Gerrard G, Jalali GR, Mehta A, et al. Cytogenetics of multiple myeloma: interpretation of fluorescence in situ hybridization results. Br J Haematol. 2003;120:944–52.
Elnenaei MO, Hamoudi RA, Swansbury J, Gruszka-Westwood AM, Brito-Babapulle V, Matutes E, et al. Delineation of the minimal region of loss at 13q14 in multiple myeloma. Genes Chromosomes Cancer. 2003;36:99–106.
Lu W, Takahashi H, Furusato M, Maekawa S, Nakano M, Meng C, et al. Allelotyping analysis at chromosome 13q of high-grade prostatic intraepithelial neoplasia and clinically insignificant and significant prostate cancers. Prostate. 2006;66:405–12.
Rowntree C, Duke V, Panayiotidis P, Kotsi P, Palmisano GL, Hoffbrand AV, et al. Deletion analysis of chromosome 13q14.3 and characterisation of an alternative splice form of LEU1 in B cell chronic lymphocytic leukaemia. Leukemia. 2002;16:1267–75.
Tyybakinoja A, Vilpo J, Knuutila S. High-resolution oligonucleotide array-CGH pinpoints genes involved in cryptic losses in chronic lymphocytic leukaemia. Cytogenet Genome Res. 2007;118:8–12.
Kapanadze B, Kashuba V, Baranova A, Rasool O, van Everdink W, Liu Y, et al. A cosmid and cDNA fine physical map of a human chromosome 13q14 region frequently lost in B-cell chronic lymphocytic leukaemia and identification of a new putative tumor suppressor gene, Leu5. FEBS Lett. 1998;426:266–70.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukaemia. Proc Natl Acad Sci USA. 2002;99:15524–9.
Baranova A, Hammarsund M, Ivanov D, Skoblov M, Sangfelt O, Corcoran M, et al. Distinct organization of the candidate tumor suppressor gene RFP2 in human and mouse: multiple mRNA isoforms in both species- and human-specific antisense transcript RFP2OS. Gene. 2003;321:103–12.
Wolf S, Mertens D, Schaffner C, Korz C, Korz C, Döhner H, et al. B-cell neoplasia associated gene with multiple splicing (BCMS): the candidate B-CLL gene on 13q14 comprises more than 560 kb covering all critical regions. Hum Mol Genet. 2001;10:1275–85.
van Everdink WJ, Baranova A, Lummen C, Tyazhelova T, Looman MW, Ivanov D, et al. RFP2, c13ORF1, and FAM10A4 are the most likely tumor suppressor gene candidates for B-cell chronic lymphocytic leukaemia. Cancer Genet Cytogenet. 2003;146:48–57.
Mertens D, Wolf S, Schroeter P, Schaffner C, Döhner H, Stilgenbauer S, et al. Down-regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B-cell chronic lymphocytic leukaemia. Blood. 2002;99:4116–21.
Ivanov DV, Tyazhelova TV, Lemonnier L, Kononenko N, Pestova AA, Nikitin EA, et al. A new human gene KCNRG encoding potassium channel regulating protein is a cancer suppressor gene candidate located in 13q14.3. FEBS Lett. 2003;539:156–60.
Rybalchenko V, Prevarskaya N, Van Coppenolle F, Legrand G, Lemonnier L, Le Bourhis X, et al. Verapamil inhibits proliferation of LNCaP human prostate cancer cells influencing K+ channel gating. Mol Pharmacol. 2001;59:1376–87.
Lewis RS, Cahalan MD. Potassium and calcium channels in lymphocytes. Annu Rev Immunol. 1995;13:623–53.
Smith GA, Tsui HW, Newell EW, Jiang X, Zhu XP, Tsui FW, et al. Functional up-regulation of HERG K+ channels in neoplastic hematopoietic cells. J Biol Chem. 2002;277:18528–34.
Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989.
Kim YH, Proust JJ, Buchholz MJ, Chrest FJ, Nordin AA. Expression of the murine homologue of the cell cycle control protein p34cdc2 in T lymphocytes. J Immunol. 1992;149:17–23.
Grace MB, McLeland CB, Gagliardi SJ, Smith JM, Jackson WE, Blakely WF. Quantitative RT-PCR assay using simultaneous measurement of four fluorophores for radiation-responsive gene targets. Clin Chem. 2003;49:1467–75.
Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Pontén F, et al. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18:1670–82.
Skoblov M, Shakhbazov K, Oshchepkov D, Ivanov D, Guskova A, Ivanov D, et al. Human RFP2 gene promoter: unique structure and unusual strength. Biochem Biophys Res Commun. 2006;342:859–66.
Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82.
Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–86.
Cho YG, Kim CJ, Song JH, Rhie DJ, Park YK, Kim SY, et al. Genetic and expression analysis of the KCNRG gene in hepatocellular carcinomas. Exp Mol Med. 2006;38:247–55.
Liu JP, Liu NS, Yuan HY, Guo Q, Lu H, Li YY. Human homologue of SETA binding protein 1 interacts with cathepsin B and participates in TNF-Induced apoptosis in ovarian cancer cells. Mol Cell Biochem. 2006;292:189–95.
Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S, et al. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci. 2005;25:8338–46.
Suehara Y, Kondo T, Seki K, Shibata T, Fujii K, Gotoh M, et al. Pfetin as a prognostic biomarker of gastrointestinal stromal tumors revealed by proteomics. Clin Cancer Res. 2008;14:1707–17.
Acknowledgements
The authors are grateful to Andy Patamawenu and Dr. Manpreet Randhawa for the help with FACS analysis, Amy VanMeter for an inclusion of our samples in the RPA assay, Stephanie Schmidt for cell culture, Masoomeh Sikaroodi and Dr. Pat Gillevet for sequencing, Dr. Dan Cox for the help with cell imaging, and Dr. Chethana Manjunath for phenotype descriptions of the cell lines. This research was performed in frame of NIH R1R15CA113331-01 “KCNRG gene as candidate tumor suppressor for CLL and MM” (2005–2009), RFFI 07-04-00379-a, 07-04-12232-ofi, and 04-04-08154-ofi.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Comparative analysis of the RFP2/KCNRG locus in human, rat, and mouse genomes. MAR/SARs are denoted by blue squares. Mouse RFP2/KCNRG locus does not possess any MAR/SAR elements (PDF 52 kb)
Supplementary Figure 2
A phylogenetic tree of prealigned T1 domains of KCNRG-like proteins (PDF 15 kb)
Supplementary Table 1
The list of the phosphoproteome endpoints assessed by reverse-phase phosphoproteome analysis (XLS 33 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Birerdinc, A., Nohelty, E., Marakhonov, A. et al. Pro-apoptotic and antiproliferative activity of human KCNRG, a putative tumor suppressor in 13q14 region. Tumor Biol. 31, 33–45 (2010). https://doi.org/10.1007/s13277-009-0005-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-009-0005-0