Abstract
The Diatrypaceae occur worldwide and comprise a number of pathogens of woody crops, forest and ornamental tree species. Despite the taxonomic difficulties within this family, interest in the Diatrypaceae has increased recently, mainly due to the recent detection of these fungi in the premium grape growing regions of California. In the present study, we investigated the diversity and host range of diatrypaceous fungi from prominent wine grape growing regions in South Australia, New South Wales and Western Australia. Approximately 100 isolates were collected from grapevine and other woody plants and compared with reference collections from the United States and Europe. Phylogenetic analyses of the complete sequence of the internal transcribed spacer (ITS) region of the ribosomal DNA and partial sequence of the β-tubulin gene, combined with morphological analyses separated 12 species. These included the previously described species Cryptovalsa ampelina, C. rabenhorstii, Diatrype brunneospora, Eutypa lata, E. leptoplaca, Eutypella australiensis, E. citricola, a Cryptosphaeria sp. and a Diatrype sp., whereas Diatrypella vulgaris, Eutypella cryptovalsoidea and E. microtheca are described as new. Seven species were isolated from grapevine but the prevalence of Diatrypaceae in grapevine cankers varied among the regions surveyed. In many instances in WA and NSW, these newly reported fungi were more widespread and abundant than E. lata. This study provides new information to assist with diagnosis of the causal agents of dieback and canker diseases in Australia and development of management strategies. Further studies to characterize the pathogenicity of diatrypaceous species to grapevines and to elucidate the biology of these fungi are underway.
Similar content being viewed by others
Introduction
Species of Diatrypaceae (Xylariales) are widespread inhabitants of dead wood and bark of a broad variety of plants around the world. Principal morphological characteristics of Diatrypaceae consist of perithecial ascomata embedded usually in a black-colored stroma, long stalked asci and allantoid ascospores (Glawe and Rogers 1984; Rappaz 1987). The current edition of Dictionary of the Fungi reports 13 genera and more than 220 morphological species (Kirk et al. 2008), with the most common genera comprising Cryptosphaeria Ces. & De Not., Cryptovalsa (Ces. & De Not.), Diatrype Fr., Diatrypella (Ces. & De Not.) De Not., Eutypa Tul. & C. Tul., and Eutypella (Nitschke) Sacc.
While several species, such as Cryptovalsa ampelina (Nitschke) Fuckel, Eutypa lata (Pers.: Fr.) Tul. & C. Tul. and E. leptoplaca (Mont.) Rappaz, are cosmopolitan (Carter 1991; Trouillas and Gubler 2004; Trouillas et al. 2010a, b), others, most notably Diatrype disciformis (Hoffm. : Fr.) Fr. are thought be extremely rare outside Europe (Rappaz 1987). Furthermore, some species appear to be associated with a specific host, for instance Eutypa maura (Fr. : Fr.) Fuckel on Acer pseudoplatanus (Rappaz 1987), while others, specifically E. lata, E. leptoplaca and C. ampelina demonstrate wider host ranges (Carter et al. 1983; Rappaz 1987; Trouillas and Gubler 2004; Trouillas and Gubler 2010; Trouillas et al. 2010a, b). Regardless, species within the Diatrypaceae have, for the most part, been considered saprotrophic, although some species appear to be especially well established in the wood of recently dead host plants (Tiffany and Gilman 1965). Nevertheless, a few species in this family are known as severe plant pathogens of woody crops, landscape and forest trees in the United States (US) and Europe (Carter 1957; Carter 1991; Davidson and Lorenz 1938; Hinds and Laurent 1978; Hinds 1981; Moller and Kasimatis 1978; Munkvold and Marois 1994; Sinclair and Lyon 2005; Jurc et al. 2006). Among those of economical importance, E. lata has been studied extensively both in Australia and around the world as the causal agent of Eutypa dieback of grapevine (Vitis vinifera L.) and apricot (Prunus armeniaca L.) (Carter 1957; Carter 1991).
The biodegradation potential of diatrypaceous strains was recently investigated (Pildain et al. 2005). This study has shown that some members of the Diatrypaceae family produce cellulase and lignin-degrading enzymes, extracellular enzymes that catalyse the hydrolysis of cellulose and breakdown of lignin in the cell walls of plants, thus affording some species the physiological capacity to produce wood decay (Pildain et al. 2005). Recent studies in the US reported several species as putative pathogens of grapevine (Rolshausen et al. 2004; Catal et al. 2007; Trouillas and Gubler 2004; Trouillas and Gubler 2010; Trouillas et al. 2010a, b; Úrbez-Torres et al. 2009). Eutypella vitis (Schwein.:Fr.) Ellis and Everh. [syn.: E. aequilinearis (Schwein.:Fr.) Starb.] and Diatrypella sp. were shown to be somewhat pathogenic to grapevine in Texas (Úrbez-Torres et al. 2009). In California, E. leptoplaca, Diatrype stigma (Hoffm. : Fr.) Fr., D. whitmanensis J.D. Rogers & Glawe, Cryptosphaeria pullmanensis Glawe and C. ampelina were shown to infect grapevine wood, causing decay of vascular tissues (Trouillas and Gubler 2004; Trouillas and Gubler 2010). Californian studies also suggested a possible correlation between grapevine infection by diatrypaceous species and the immediacy of fungal fruiting bodies (perithecia) on natural and ornamental host plants in the surrounding ecosystems, often in close proximity to vineyards (Trouillas and Gubler 2010; Trouillas et al. 2010a, b).
Berlese (1900) introduced the earliest large-scale taxonomic study of Diatrypaceae, providing excellent illustrations for many species. Rappaz (1987) revised the family examining thoroughly original descriptions and types around the world. To date, his work provides the most comprehensive treatment on the taxonomy of octosporous Diatrypaceae. In North America, Ellis and Everharts (1892) proposed descriptions for numerous Diatrypaceae, including polysporous genera. Later, Tiffany and Gilman (1965), and Glawe and Rogers (1984), described Diatrypaceae from Iowa and from the Pacific Northwest, respectively. Lately, Vasilyeva and Stephenson (2004, 2005, 2006, 2009) described several species from the Great Smoky Mountains National Park in the eastern US, Arkansas and Texas. Additional studies have investigated the diversity of Diatrypaceae in Argentina, describing new species and new records (Romero and Carmarán 2003; Carmarán et al. 2009).
The current generic delineation and classification of Diatrypaceae as proposed by Rappaz (1987) is based primarily on characters of the teleomorphic states, including stroma morphology and organization of perithecia. However, much overlap of these taxonomic features exists among the current diatrypaceous genera. For example, the concept of Diatrype as delimited by Rappaz (1987) has, in some instances, no clear separation from either Eutypa or Eutypella (Vasilyeva and Stephenson 2004). Overall, the taxonomy of the Diatrypaceae is outdated making the identification of these fungi particularly difficult. Published diagnoses for these species are often vague and incomplete, while most original descriptions as well as types are largely inaccessible or lost. The current classification of diatrypaceous genera remains provisional and there is an urgent need to revise the classification of the family and test the significance of generic concepts using molecular phylogeny. Preliminary attempts at phylogenetic classification using molecular data as well as morphological characters remained inconclusive regarding the evolutionary relationships of these fungi (Acero et al. 2004; Carmarán et al. 2006; Trouillas et al. 2010a, b).
In Australia, little work has been conducted to investigate the diversity and taxonomy of diatrypaceous fungi. Most studies have focused on the apricot and grapevine pathogen E. lata, which is widespread across South Australian (SA) vineyards (Carter 1991; Highet and Wicks 1998; Lardner et al. 2005; Sosnowski et al. 2007). However, a number of additional species were documented more recently. In 2004, Mostert et al. (2004) accounted for the occurrence of C. ampelina from vineyards both in the Coonawarra and Eden Valley regions of SA and went on demonstrate that this fungus could cause disease in grapevine shoots. Additional Diatrypaceae were also reported from surveys of fungi associated with canker diseases in grapevine in New South Wales (NSW), but identification of these isolates remained incomplete (Pitt et al. 2010).
Diatrypaceous fungi from native plant species have been reported sporadically in Australia. In his handbook of “Australian fungi”, Cooke (1892) described seven putative species of Diatrypaceae, including Diatrype glomeraria Berk, Diatrype stigma, Diatrype chlorosarca Berk. & Broome, Cryptovalsa elevata Berk., E. lata, E. lubidunda (Sacc.) Thüm. (= E. leprosa [Pers.] Berl.), and Eutypella stellulata (Fr. : Fr.) Sacc. Additional species were described from intertidal host plants in north Queensland, including Cryptovalsa halosarceicola K.D. Hyde on Halosarcia halocnemoides (Nees) Paul G. Wilson in a mangrove at Cairns Airport (Hyde 1993), Eutypa bathurstensis K.D. Hyde & Rappaz (Hyde and Rappaz 1993) and Eutypella naqsii K.D. Hyde (Hyde 1995) on Avicennia sp. at Bathurst Heads. Later, Yuan (1996) documented Cryptovalsa protracta (Pers.) De Not., Diatrype stigma and Eutypella scoparia (Schwein. : Fr.) Ellis & Everh. on Acacia and Eucalyptus plants on Melville Island in the Northern Territory, while Trouillas et al. (2010a, b) described two additional species from native Acacia shrubs in the Coorong National Park, SA. To the best of our knowledge, the above references constitute the only studies that illustrate the diatrypaceous mycota in Australia.
During this study, we conducted surveys and investigated the diversity of diatrypaceous fungi associated with grapevines and other woody plants and in SA, NSW and Western Australia (WA). In many instances, fungal colonies displaying morphological characteristics typical of Diatrypaceae were isolated from diseased grapevines. Fruiting bodies typical of Diatrypaceae were also observed from grapevines. The diversity, identity and distribution of these fungi in the main wine grape growing regions of Australia are currently unknown. Hence, much work is necessary not only in the collection and identification of the various species, but also in the determination of their pathogenicity to grapevines and role in the overall complex of grapevine canker diseases. The objectives of this study were to collect, identify and describe the diatrypaceous fungi in and near Australian vineyards, and characterize species using morphology and molecular phylogeny.
Materials and methods
Origin and deposit of isolates
During spring and summer of 2008 and 2009, we obtained strains of Diatrypaceae from cankers in infected grapevine spurs, cordons or trunks, and from fruiting bodies on dead grapevines as well as dead wood of native, ornamental and cultivated plants neighboring vineyards. Viticultural regions sampled included the Barossa Valley, Adelaide Hills, McLaren Flat, and Coonawarra in SA; the Hunter Valley, the Riverina, and Southern NSW regions; and the Swan Valley, Margaret River, and Great Southern regions of WA. Isolates from the National Wine and Grape Industry Centre (Charles Sturt University, Wagga Wagga, NSW, Australia) collected in previous surveys (Pitt et al. 2010) were also used in this study. The geographic origin and host range of the specimens collected during this study are summarized in Table 1.
Isolates were grown from ascospores or from hyphae in infected grapevine wood as described by Trouillas et al. (2010a, b). Pure cultures were obtained by transferring single hyphal tips onto potato dextrose agar (PDA; Oxoid Ltd, Basingstoke, Hampshire, England) amended with 100 ppm tetracycline (PDA-tet). Representative isolates, including ex-type cultures (fresh cultures) of Diatrypaceae from Australia were deposited both at Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands (accession no: CBS128327- CBS128339), and at the Australian Scientific Collections (DAR), Industry & Investment NSW, Orange, NSW, Australia (accession no: DAR81030-DAR81042). Dry specimens (bark and/or wood) containing the perfect stage of each fungal isolate were also deposited at DAR.
Identification and morphological analysis
Fruiting bodies of Diatrypaceae were identified in conformity with the treatments of Glawe and Rogers (1984) and Rappaz (1987). In addition, putative new species of Eutypella, Diatrypella and Cryptovalsa were compared with descriptions and illustrations in Saccardo’s Sylloge Fungorum vol. 1 (1882), Ellis and Everharts (1892), and Berlese (1900) to verify species originality. Specimens from Australia were also compared with reference specimens from California (Trouillas et al. 2010a, b) using morphological and phylogenetic analyses. Microscopic examinations were carried out with standard light microscopy on an Olympus Provis AX70TRF (Olympus Optical Co. Ltd., Japan) microscope fitted with a ColorView IIIu digital camera (Soft Imaging Systems (SIS) GmbH, Munster, Germany). Conidial masses as well as perithecial contents were mounted in water and observed by brightfield microscopy. Digital images were recorded using analySIS LS Research 2.41 (SIS) and measurements for at least 30 conidia, 50 ascospores and 20 asci were made for each specimen. To study colony morphology and conidial production, cultures on PDA were maintained in incubators under controlled conditions of intermittent fluorescent lighting (12 h) at 24°C.
DNA isolation, amplification and phylogenetic analyses
DNA extractions were performed as described by Pitt et al. (2010). Total genomic DNA was extracted from pure cultures after transferring colonized agar plugs into 50 mL Falcon tubes filled with 20 mL of potato dextrose broth (Oxoid Ltd., Basingstoke, Hampshire, England). Broth cultures were then incubated on a Sartorius Certomat BS-1 (Goettingen, Germany) orbital shaker revolving at 90 rpm for 7 days at 25°C. Mycelia were collected by filtration, lyophilized and DNA was extracted using the Qiagen Plant Mini Kit according to the manufacturer’s instructions (Qiagen Pty Ltd, Clifton Hills, Vic., Australia). The internal transcribed spacer regions (ITS1 and ITS2), including the 5.8 S rDNA operon of the nuclear ribosomal DNA region were amplified by the polymerase chain reaction (PCR) using primers ITS5 and ITS4 (White et al. 1990). Partial sequence of the β-tubulin gene was amplified using primers Bt2a and Bt2b (Glass and Donaldson 1995). Each PCR tube contained 0.1 volume of 10× buffer (15 mM MgCl2, Qiagen), 200 mM each of dNTPs, 0.15 mM of each primer, 1 unit of HotStar Taq DNA polymerase (Qiagen), and ~50 ng of DNA template, and were adjusted with sterile nanopure water to a total volume of 50 μL. PCR was performed using an Eppendorf Master Thermocycler (Hamburg, Germany). Amplification was accomplished by an initial step of 2 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 58°C, and 1.5 min at 72°C, with a final extension of 5 min at 72°C. PCR products were separated by electrophoresis on 1% agarose gels containing 0.5× Tris-borate-EDTA buffer. Positive amplifications were confirmed by photography under UV light following staining with ethidium bromide (0.5 mg/L). PCR products were purified using the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA). Both strands of the ITS and β-tubulin regions were sequenced by the Australian Genome Research Facility (University of Queensland, St Lucia, Qld, Australia). Sequencing results were edited and assembled using Sequencher™ version 3.1.1. Sequences were aligned using ClustalW multiple alignment program (Thompson et al. 1994) and were adjusted manually using BioEdit Sequence Alignment Editor Version 7.0.8. (Hall 1999). Phylogenetic analyses were performed with PAUP version 4.0b10 (Swofford 1999) using maximum parsimony (MP) with a heuristic search and 1000 random addition sequence replicates. Tree bisection-reconnection (TBR) was used as the branch swapping algorithm. Branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. Ambiguously aligned regions were not excluded for pyhlogenetic analyses and alignment gaps were treated as missing data. Measures including tree length, consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated. Bootstrap support (BS) was calculated using 1000 replicates to test branch strength. Sequences have been deposited into GenBank (HQ692458-HQ692622). To accelerate the process, phylogenetic analyses were run using a single representative of each haplotype. Sequences of Xylaria hypoxylon, Daldinia concentrica, Anthostomella eucalytorum, A. protea, Nemania aenea and Camilea tinctor from GenBank were used as outgroup in the ITS analysis. Beta tubulin trees were rooted using E. scoparia as outgroup.
Results
Phylogenetic analyses
ITS and β-tubulin sequences were obtained for approximately 90 isolates of Diatrypaceae collected in Australia. Unique ITS sequences or haplotypes were aligned with approximately 50 GenBank reference sequences, while the β-tubulin dataset included 24 sequences obtained from GenBank.
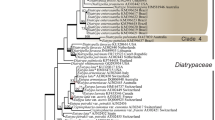
The ITS analysis comprised 74 taxa and 636 characters, of which 276 were constant, 83 parsimony-uninformative and 277 parsimony-informative. The heuristic search using the ITS dataset resulted in 36 most parsimonious trees of similar topologies, each comprising 1518 steps (CI = 0.4302, RI = 0.7444, RC = 0.3202 and HI = 0.6126). One of the 36 most parsimonious (MP) trees is shown in Fig. 1.
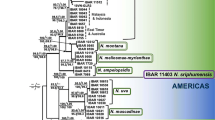
In contrast, the β-tubulin dataset contained 45 taxa and 417 characters, of which 207 were constant, 17 parsimony-uninformative, and 194 parsimony-informative. The MP analysis resulted in 10 trees, each with a length of 703 steps (CI = 0.5391, RI = 0.8253, RC = 0.4450 and HI = 0.4723). Each most parsimonious tree shared the same overall topology, one of which is shown in Fig. 2.
Grouping of genera and species was generally similar for the ITS and β-tubulin analyses. Bootstrap values from the ITS and β-tubulin data sets (98% and 87% respectively) supported the occurrence of a main clade comprising several Eutypella and Cryptovalsa-like spp. (Figs. 1 – 2). E. microtheca (with 8-spored asci) grouped with the polysporous spp. Eutypella cryptovalsoidea and C. rabenhorstii (96% and 98% respectively) (Figs.1 – 2). Similarly, the octosporous D. oregonensis was closely related to various polysporous Diatrypella spp. (85% and 96% respectively) (Figs. 1 – 2).
In the ITS analysis, Diatrype spilomea, D. bullata, D. disciformis, D. stigma, D. undulata and Diatrypella quercina formed another clade and demonstrated strong affinities (96% bootstrap value) (Fig. 1 ). Eutypa spp. separated into two major clades. The first clade included E. lata, E. lata var. aceri, E. laevata, E. petrakii var. petrakii and also included C. eunomia (80% bootstrap value). The second clade included all remaining Eutypa species that were tested (94% bootstrap value) and also included E. prunastri and D. polycocca (Fig. 1). Isolates NSW01PO−NSW04PO appeared to be closely related to C. lignyota.
Taxonomy
Descriptions are provided for novel or unusual species. Tables 2 and 3 illustrate conidial, ascus and ascospore sizes for all isolates examined in this study. Measurements under the following descriptions represent averaged sizes obtained from the different isolates.
Cryptovalsa rabenhorstii (Nitschke) Sacc., Myc. Ven. 135, tab. XIV.
(Fig. 3)
Morphology of Cryptovalsa rabenhorstii. a. Perithecial stroma in the bark of a lignified cane of Vitis vinifera; b. Emerging perithecial ostioles surrounded with white ectostroma and perithecial cavities; c. Long-pedicellate polysporus ascus; d. Mature (light brown) and immature (hyaline) ascospores; e. Colony after 29 days on 85 mm diam PDA dish incubated under intermittent fluorescent lighting (12 h). Bars = 1 cm in a; 1 mm in b; 50 μm in c–d
Basionym: Valsa rabenhorstii Nitschke Pyr. Germ.
Synonym: Sphaeria spiculosa var. robiniae Rabenh., in Exsicc. Klotzsch, Herb. myc.
Stromata in bark of lignified canes (V. vinifera), poorly developed, perithecia buried in the inner bark and scattered in subvalsiform groups of 2–3, or fairly irregularly in larger groups, raising the epidermis which is not discolored and remains attached, or which rupture longitudinally revealing groups of black ostioles occasionally sheltered around a white ectostroma, which apparently facilitate pressuring and splitting of the bark; perithecia outer surface coated with white, powdery entostroma, 0.35–0.55 mm diam, ostioles poorly emerging, more or less distinctly quadrisulcate. Asci long-pedicellate, polysporous, p. sp. (55−)70−90(−95) × (15−)18−22(−27) μm. Ascospores hyaline when immature turning yellowish to light-brown at maturity, sub-allaintoid, cylindrical to oblong, (10−)13.5−15(−17.5) × (3.2−)4−5(−6) μm. Colonies white with rather irregular margin. Conidia not seen.
Hosts. Vitis vinifera (Australia, WA), Sambuscus nigra (USA, CA).
Notes This species has characteristics typical of members of the genus Cryptovalsa, and resembles closely descriptions of C. rabenhorstii (Nitschke 1867; Saccardo 1882) as well as the illustration by Berlese (1900) of C. ampelina, C. rabenhorstii var. rosarum and C. rabenhorstii var. eutypelloidea. However, as we could not find the type specimen nor obtain culture collections for this species, identification remains tentative. Also, phylogenetic analyses show affinities of this fungus with Eutypella spp. The assignment of this isolate to the genus Cryptovalsa may therefore require future reconsideration. Hence, it is preferable not to propose a novel combination for this species until identification of types and further large scale phylogenetic studies of the Diatrypaceae can be conducted.
Specimens examined. AUSTRALIA, WA, Great Southern regions, on lignified canes of Vitis vinifera on the ground, Nov. 2009, F. P. Trouillas, coll. number WA07CO, DAR81041, CBS128338; and coll. number WA08CB, DAR81042, CBS128339.
Diatrypella vulgaris Trouillas, W. M. Pitt & Gubler, sp. nov.
(Fig. 4)
Morphology of Diatrypella vulgaris. a. Pustulate stromata with white entostroma embedded in the bark of Fraxinus angustifolia; b. Group of polysporus asci and paraphyses; c. Close up on the rather short-stalked ascus, with wide and lengthy spore-bearing portion; d. Colony after one month incubation in the dark at 25°C on 85 mm PDA dish; e. Allantoid ascospores. Bars = 1 mm in a; 50 μm in b–c; 50 μm in e
MycoBank: MB 519404
Etymology Vulgaris, meaning ordinary, to account for the typical Diatrypella morphology of this fungus.
Stromata erumpentia, in pustulis 1–4 μm longis, saepe a nigro lineamento in infero ligno evidente circumscripta, per corticem vel lignum dehiscentia atque a reliqua adhaerente cute vel ligneis fragmentis saepe circumfusa, incomposita et congruente vel hemispherica atque iuxta ligneis striis oblonga formis variantia. Perithecia circinata vel ovoidea, aliquando compressa, ex albo entostroma amplexa, 0.25–0.45 mm diametro. Ostiola sulcata, parum eminentia. Asci brevioribus caulis, paraphysati, polyspori, parte sporifera (65−)80−130(−155) × (12−)18–20 μm. Ascospores allantoideae, corpore flavidae (7−)8−10(−12) × 2–2.5 μm. Albae coloniae leviter fuscae aetate se vertentes, una specie cum subexcelso mycelio pycnidia constituente, conidia ad parum lutea corpora manantia. Conidia fili instar, 25–40(−55) × (1−)1.5–2 μm.
Stromata well developed, in pustules 1–4 mm in length, often delimited with a black line perceptible in the wood below, bursting through bark or wood and often surrounded by remaining adherent epidermis or wood fragments, varying in shape from irregular and confluent to hemispherical and oblong following wood striations, perithecia circular to ovoid, occasionally compressed, surrounded by white entostroma, 0.25–0.45 mm diam, ostioles sulcate, only slightly prominent. Asci with moderately short stalks, paraphysate, polysporous, p. sp. (65−)80−130(−155) × (12−)18–20 μm. Ascospores allantoid, yellowish in mass (7−)8−10(−12) × 2–2.5 μm. Colonies white becoming light brown with age, homogeneous with rather moderate aerial mycelium, forming pycnidia exuding conidia in light orange masses. Conidia filiform, 25–40(−55) × (1−)1.5–2 μm.
Hosts. Citrus paradisi, Fraxinus angustifolia, Schinus molle var. areira (Australia, NSW).
Notes. This fungus shows morphological characteristics typical of fungi in the genus Diatrypella and resembles in many aspects earlier descriptions of D. verruciformis and D. pulvinata. However, this species can be distinguished on characteristics of the asci which are longer and unusually wide, and which bear longer ascospores than most previously described species (commonly 6–8 μm) (Saccardo 1882; Ellis and Everharts 1892; Berlese 1900; Glawe and Rogers 1984). Also, ITS sequences of this fungus differed from all Diatrypella spp. sequences available in GenBank, including D. pulvinata and D. verruciformis.
Specimens examined. AUSTRALIA, NSW, Hunter Valley, on dead branches of Citrus paradisi, Dec. 2008, HOLOTYPE: F. P. Trouillas & W. M. Pitt, coll. number HVGRF03, DAR81030, CBS128327; on dead branches of Fraxinus angustifolia, Dec. 2008, ISOTYPE: F. P. Trouillas & W. M. Pitt, coll. number HVFRA04, DAR81031, CBS128328; on dead branches of Schinus molle var. areira, Dec. 2008, ISOTYPE: F. P. Trouillas & W. M. Pitt, coll. number HVPT01, DAR81032, CBS128329.
Eutypella citricola Speg., Anales del Museo Nacional de Buenos Aires 6: 245, 1898. (Fig. 5)
Morphology of Eutypella citricola. a. Pustulate stromata aggregated in the bark of Citrus sinensis; b. Pustulate stromata on lignified canes of Vitis vinifera; c. Long-stalked ascus; d. Allantoid ascospores; e. Colony after one month incubation in the dark at 25°C on 85 mm PDA dish. Bars = 1 mm in a; 5 mm in b; 50 μm in c; 20 μm in d
Stromata in the bark or wood, which appear rugous, in pustules scattered or aggregated into large surface, pustules often delineated with black line; perithecia surrounded by white, powdery entostroma, attached onto the outer surface, circular to ovoid, sometimes compressed with others, 0.25–0.5 mm diam; ostioles raising and piercing the periderm; ostioles slightly emerging through the periderm, in contact within small groups, well define, 3–4 sulcate. Asci 8-spored, clavate, p. sp. 55–80 × 7.5–9 μm. Ascospores allantoid, subhyaline to light yellow, (9−)10.5–12(−13) × 2–3 μm. Colonies white and even, moderate aerial mycelium, forming numerous dots of melanized mycelium spread across the media and visible from the underside after 30 days incubation on PDA at 24°C. Conidia filiform 15–20(−25) × 1.5–2 μm.
Hosts Citrus limon, Citrus sinensis, Citrus paradisi (Australia, NSW) ; Schinus molle var. areira, Ulmus procera (Australia, SA) ; Vitis vinifera (Australia, NSW ; USA, CA).
Notes Collections from Australia were morphologically identical to the type specimen of Eutypella citricola and the identification of these isolates is confident.
Specimens examined ARGENTINA, La Trinidad, prov. Tucumán, on branch of Citrus aurantium, Jan. 1895, HOLOTYPE: Speg., LPS-2120. AUSTRALIA, NSW, Hunter Valley, on dead branches of Vitis vinifera, Dec. 2008, F. P. Trouillas & W. M. Pitt, coll. number HVVIT07, DAR81033, CBS128330; on dead branches of Citrus sinensis, Dec. 2008, F. P. Trouillas & W. M. Pitt, coll. number HVOT01, DAR81034, CBS128331; on dead branches of Citrus paradisi, Dec. 2008, F. P. Trouillas & W. M. Pitt, coll. number HVGRF01, DAR81037, CBS128334; WA, Swan Valley, on dead branches of Citrus limon, Nov. 2009, F. P. Trouillas, coll. number WA04LE, DAR81035, CBS128332; on dead branches of Vitis vinifera, Nov. 2009, F. P. Trouillas, coll. number WA05SV, DAR81036, CBS128333; SA, Adelaide, Waite Campus, on dead branches of Ulmus procera, Nov. 2008, F. P. Trouillas, coll. number ADEL100; on dead branches of Schinus molle var. areira, Nov. 2008, F. P. Trouillas, coll. number ADSC100.
Eutypella cryptovalsoidea Trouillas, W. M. Pitt & Gubler, sp. nov.
(Fig. 6)
MycoBank: MB 519406
Etymology. Cryptovalsoidea, referring to the morphological similitude of this fungus with Cryptovalsa.
Stromata plerumque in cortice, male evoluta circa fundum perithecialem, nigra, effusa atque paulo callosiora circa cervices peritheciales sub peridermio. Perithecia plus minus inter se coniuncta et ad copiosos coetus congruentia, inaequabiliter constratos. Ostiola hemisphaerica, saepe perforata, singula vel coniunctim per corticem eminentia. Asci clavati vel fusiformes, longe pedicellati, polyspori, parte sporifera 65–120 × 15–20 μm. Ascosporae flavidae, in corpore aquiliorae, allantoideae vel sub-allantiodeae, 8–12(−13.5) × 2–3 μm. Coloniae albae cum subexcelso mycelio tenuique areo-roseo inferiore. Conidia non evidentia.
Stromata mostly in bark, poorly developed around the perithecial base, black, effuse and rather crusty around perithecial necks below the periderm; perithecia more or less in contact and confluent into large groups, irregularly scattered; ostioles hemispherical, often perforated, emerging singly or in groups through bark. Asci clavate to spindle-shape, long-pedicellate, polysporous, p. sp. 65–120 × 15–20 μm. Ascospores yellowish, darker in mass, allaintoid to sub-allaintoid, 8–12(−13.5) × 2–3 μm. Colonies white with rather moderate aerial mycelium and slight orange-pink underside. Conidia not seen.
Hosts. Ficus carica (Australia, NSW).
Notes. The present species displays some features of morphology typical of Cryptovalsa (poorly developed stroma, polysporous ascus) as well as Eutypella (perithecial necks erumpent in groups). Because of the polyporous ascus, this species could be referred as Cryptovalsa under the current classification scheme for Diatrypaceae. However, size and shape of the polysporous asci differed from all Cryptovalsa species previously described from Ficus carica and additional host plants. (Saccardo 1882; 1905; 1926; Berlese 1900; Spooner 1981).
Specimens examined. AUSTRALIA, NSW, Hunter Valley, on dead branches of Ficus carica, Dec. 2008, HOLOTYPE: F. P. Trouillas & W. M. Pitt, coll. number HVFIG02, DAR81038, CBS128335.
Eutypella microtheca Trouillas, W. M. Pitt & Gubler, sp. nov.
(Fig. 7)
Morphology of Eutypella microtheca. a. Stromata in bark of Citrus paradisi elevating the periderm surface and minute perithecial cavities; b. Long-stalked ascus; c. Allantoid ascospores; d. Pink underside of colony after 5 days on 85 mm diam PDA dish incubated under intermittent fluorescent lighting (12 h); e. Light pink colony with cottony mycelium aggregates after one month incubation in the dark at 25°C on 85 mm PDA dish. Bars = 1 mm in a; 50 μm in b; 50 μm in c
MycoBank: MB 519407
Etymology. Microtheca, referring to the small diam of the perithecia.
Stromata in cortice: subrigentia extremitatem peridermii (tumens aspectus), quod ex eminentibus, non-proiectis ostiolis laceratur; stromata in ligno: paulo magis eutypeia, ligni superficiem fuscantia tollentiaque. Perithecia plus minusve inter se coniuncta, globosa vel ovoidea, a candida, pulverulenta trama circumfusa, minuta, 0.15–0.25(−0.3) μm diametro; ostiola 3–4 sulcata. Asci octospori, clavati, longe stipitati, parte sporifera 35–55(−60) × 7–9 μm. Ascosporae allantoideae, subhyalinae vel flavescentes, 8–10(−11) × 2–2.5 μm. Coloniae roseae, ad canum vergentes et crebra pycnidia conficientes. Conidia fili instar 16–22(−25) × 1.5–2 μm.
Stromata in bark: elevating the periderm surface (swollen appearance), which become ripped off by the emerging, non-prominent ostioles; stromata in wood: rather eutypoid, blackening and raising the wood surface. Perithecia more or less in contact, round to ovoid, surrounded by white, powdery entostromatic tissue, minute, 0.15–0.25(−0.3) mm diam; ostioles 3–4 sulcate. Asci 8-spored, clavate, long-stipitate, p. sp. 35–55(−60) × 7–9 μm. Ascospores allantoid, subhyaline to light yellow, 8–10(−11) × 2−2.5 μm. Colonies light pink, turning grey and forming numerous pycnidia with age. Conidia filiform 16−22(−25) × 1.5−2 μm.
Hosts. Citrus paradisi (Australia, NSW), Vitis vinifera (Australia, NSW; USA, CA), Ulmus procera (Australia, SA).
Notes. This fungus differs from all Eutypella species recognized by Rappaz (1987) mostly due to its smaller perithecia (commonly <250 μm). This fungus is also distinctive as a result of the light pink coloration of colonies when grown on PDA and PDA-tet.
Specimens examined. AUSTRALIA, NSW, Hunter Valley, on dead branches of Citrus paradisi, Dec. 2008, HOLOTYPE: F. P. Trouillas, coll. number HVGRF02, DAR81039, CBS128336; on dead branches of Vitis vinifera, Dec. 2008, ISOTYPE: F. P. Trouillas, coll. number HVVIT05, DAR81040, CBS128337.
Discussion
Phylogenetic analyses of both the ITS regions of the rDNA and partial sequence of the β-tubulin gene identified 12 diatrypaceous species from various woody host plants in Australia (shown in bold in Figs. 1 and 2), including the recently described D. brunneospora and E. australiensis (Trouillas et al. 2010a, b). Comparison with reference sequences obtained from GenBank facilitated the identification of C. ampelina, E. leptoplaca, and a Cryptosphaeria sp. isolated from cankers on Populus spp. in NSW and closely related to Cryptosphaeria lignyota (Fr.) Auersw. All the remaining species reported from this study were identified based on morphology. E. leptoplaca is reported from Fraxinus angustifolia, Schinus molle var. areira and Populus spp., although we failed to isolate the pathogen from grapevine despite the existence of previous records from this host in California (Trouillas and Gubler 2004). The occurrence of E. lata on naturalized and ornamental plant species in close to vineyards was confirmed. Negligible variations in the sequences of studied genes were found within each species group (or clade), which regularly included isolates both from different host plants and/or continents. ITS and β-tubulin sequences from isolates of C. ampelina, C. rabenhorstii, E. lata, E. leptoplaca, Eutypella citricola and E. microtheca from Australia appeared nearly identical to their California counterparts (Trouillas et al. 2010a, b).
Surveys for diatrypaceous fungi associated with grapevines and other woody hosts in Australia allowed the isolation of original specimens of what appeared to be new species in this family. Hence, D. vulgaris, E. microtheca and E. cryptovalsoidea are described as new species in this paper. Our collections were distinguished from previously described species by their unique morphological characters. Eutypella microtheca had exceptionally small perithecia and mycelia on PDA exhibited a pink coloration when grown in culture on PDA. Diatrypella vulgaris and E. cryptovalsoidea bore unusually long asci, which were also wider than previously recorded; these features differed quite significantly from those described for recognized polysporous species in this family. Isolates WA07CO and WA08CB from grapevine were identified as C. rabenhorstii and resemble closely early descriptions of this species by Nitschke (1867) and Saccardo (1882).
This research confirmed the abundance and diversity of Diatrypaceae harbored by grapevines, as shown in a similar study in California (Trouillas et al. 2010a, b). Among the species reported in the present study, seven were isolated from grapevine wood including C. ampelina, C. rabenhorstii, Diatrype sp., D. vulgaris, E. citricola, E. lata and E. microtheca. The incidence and distribution of Diatrypaceae in grapevine cankers varied significantly among the regions surveyed but in many instances these newly reported fungi were more widespread and abundant than E. lata. Eutypa lata was thought to be the main diatrypaceous species associated with canker diseases in Australia, however, both E. microtheca and E. citricola appeared to be the more dominant species occurring in grapevine cankers in parts of the Hunter Valley (NSW), where E. lata remained elusive. Eutypella citricola was found abundantly in both NSW and WA vineyards. In most instances, its presence on grapevines could be explained by the proximity of abandoned citrus orchards and declining citrus trees bearing numerous perithecia of this fungus. Generally, species of Diatrypaceae encountered on grapevines also occurred on other agricultural host plants and ornamentals adjacent to, or in close proximity to vineyards. Furthermore, many of the species commonly found in Australian vineyards were identical to those isolated during previous surveys throughout California vineyards and therefore provided new information on the host range and possible origin of these fungi.
Each genus included in the phylogenetic analyses occurred in more than one clade across the MP trees suggesting polyphyletic origins of diatrypaceous genera. Analyses confirm the observation by Acero et al. (2004) that the current taxonomic scheme for the Diatrypaceae may not reflect the true evolutionary relationships of these fungi and limits between genera. Moreover, the present analyses did not allow the evolutionary history of Diatrypaceae to be elucidated, as bootstrap values were small at deep nodes within the various tree topologies. Increased sampling of taxa (within a monophyletic group) has been widely accepted as a means to increase the average accuracy of phylogenies (Rannala et al. 1998; Pollock et al. 2002; Zwickl and Hillis 2002; Heath et al. 2008). As the diatrypaceous mycota remains poorly investigated worldwide, particularly in tropical regions, exploring the overall diversity of these fungi may be necessary ultimately to resolve the evolutionary relationships in this family. We anticipate that much broader sampling of taxa combined with multigene phylogenies will be necessary in future studies to resolve the evolutionary relationships within this family. Until then, the assignment of newly discovered species into specific diatrypaceous genera may be provisional.
Number of spores per ascus (eight spores versus more than eight spores) has been used traditionally to delineate genera of the Diatrypaceae. Species with polysporous asci have been assigned to genera including Diatrypella and Cryptovalsa, which differed from one another mostly by the degree of stromatic tissue produced around the perithecia. Unfortunately, Rappaz did not consider polysporous Diatrypaceae in his work and no modern taxonomic treatment of polysporous Diatrypaceae is available. Moreover, many types for these genera remain out of reach while original descriptions are often inadequate to delineate and identify species. Delineating Diatrypella and Cryptovalsa, has proved challenging and species are often transferred between the two genera. Wehmeyer (1926) regarded polysporous Diatrypaceae as a distinct phylogenetic lineage. Glawe and Rogers (1984) argued that multispored species might have evolved independently and repeatedly within this family while Tiffany and Gilman (1965) placed the two names in synonymy. Diatrypella has also been considered as a polysporous counterpart of Diatrype, and Cryptovalsa as a polysporous counterpart of Eutypa (Vasilyeva and Stephenson 2005). As demonstrated by the present DNA-based phylogenies, the morphospecies Cryptovalsa and Eutypella as well as Diatrype and Diatrypella showed molecular affinities. These results suggest a lack of evolutionary significance of the polysporous ascus feature in the Diatrypaceae.
In this study diatrypaceous strains were commonly isolated from necrotic grapevine wood. Furthermore, certain species normally occurring as saprophytes on the native vegetation in California could occasionally infect wounded active grapevine wood (Trouillas et al. 2010a, b). Fungi in this family are likely to play important ecological functions and may ultimately contribute to the decay of their host plant, thereby affecting plant health and crop longevity. Biologically characterization of these fungi, in term of trophism and ecology, is required to elucidate their role in the decline of the host, particularly in pathosystems involving grapevines and the occurrence of trunk disease complexes. It is important that the pathogenicity of these species on grapevine is determined, and if necessary, management strategies for trunk diseases refined to include these species.
Aknowledgements
We acknowledge M. Priest curator of the Plant Pathology Herbarium (DAR), at Australian Scientific Collections Unit, Industry and Investment NSW, Orange, NSW, Australia. We gratefully acknowledge the curator of the Centraalbureau voor Schimmelcultures (CBS) culture collection. We also extend our profound gratitude to C.C. Carmarán, Departamento de Ciencias Biológicas, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires for providing types and original descriptions of diatrypaceous fungi from Argentina. We thank Australia’s grape growers and winemakers through their investment body the Australian Grape and Wine Research & Development Corporation for financial support.
References
Acero FJ, González V, Sánchez-Ballesteros J, Rubio V, Checa J, Bills GF, Salazar O, Platas G, Peláez F (2004) Molecular phylogenetic studies on the Diatrypaceae based on rDNA-ITS sequences. Mycologia 96:249–259
Berlese AN (1900) Icones Fungorum. Vol. 3. Sphaeriaceae: Allantosporae p. p. Patavii, 120 p., 162 pls
Carmarán CC, Romero AI, Giussani LM (2006) An approach towards a new phylogenetic classification in Diatrypaceae. Fungal Divers 23:67–87
Carmarán CC, Pildain MB, Vasilyeva LN (2009) The family Diatrypaceae (Ascomycota) in Argentina: new species and new records. Nova Hedwig 88:521–530
Carter MV (1957) Eutypa armeniacae Hansf. & Carter, sp. nov., an airborne vascular pathogen of Prunus armeniaca L. in Southern Australia. Aust J Bot 5:21–35
Carter MV (1991) The status of Eutypa lata as a pathogen. Monograph, Phytopathological Paper No 32. Commonwealth Agricultural Bureau, International Mycological Institute, UK
Carter MV, Bolay A, Rappaz F (1983) An annotated list and bibliography of Eutypa armeniacae. Rev Plant Pathol 62:251–258
Catal M, Jordan SA, Butterworth SC, Shilder AMC (2007) Detection of Eutypa lata and Eutypella vitis in grapevine by nested multiplex polymerase chain reaction. Phytopathology 97:737–747
Cooke MC (1892) Handbook of Australian fungi. Williams and Norgate, London, p 457
Davidson RW, Lorenz RC (1938) Species of Eutypella and Schizoxylon associated with cankers of maple. Phytopathology 28:733–745
Ellis JB, Everhart BM (1892) The North American Pyrenomycetes. Newfield, New Jersey, 793 pp
Glass NL, Donaldson GC (1995) Development of primer set designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Glawe DA, Rogers JD (1984) Diatrypaceae in the Pacific Northwest. Mycotaxon 20:401–460
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Heath TA, Hedtke SM, Hillis DM (2008) Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol 46:239–251
Highet A, Wicks T (1998) The incidence of Eutypa dieback in South Australian vineyards. Annual Technical Issue - 1998. The Australian Grape Grower and Winemaker 414:135–136
Hinds TE (1981) Cryptosphaeria canker and Libertella decay of aspen. Phytopathology 71:1137–1145
Hinds TE, Laurent TH (1978) Common aspen diseases found in Alaska. Plant Dis Rep 62:972–975
Hyde KD (1993) Cryptovalsa halosarceicola sp. nov. an intertidal saprotroph of Halosarceia halocnemoides. Mycol Res 97:799–800
Hyde KD (1995) Eutypella naqsii sp. nov. from intertidal Avicennia. Mycol Res 99:1462–1464
Hyde KD, Rappaz F (1993) Eutypa bathurstensis sp. nov. from intertidal Avicennia. Mycol Res 97:861–864
Jurc D, Ogris N, Slippers B, Stenlid J (2006) First report of Eutypella canker of Acer pseudoplatanus in Europe. Plant Pathol 55:577
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Dictionary of the fungi, 10th edn. CAB International, Wallingford
Lardner R, Stummer BE, Sosnowski MR, Scott ES (2005) Molecular identification and detection of Eutypa lata in grapevine. Mycol Res 109:799–808
Moller WJ, Kasimatis AN (1978) Dieback of grapevine caused by Eutypa armeniacae. Plant Dis Rep 62:254–258
Mostert L, Halleen F, Creaser ML, Crous PW (2004) Cryptovalsa ampelina, a forgotten shoot and cane pathogen of grapevines. Australas Plant Path 33:295–299
Munkvold GP, Marois JJ (1994) Eutypa dieback of sweet cherry and occurrence of Eutypa lata perithecia in the central valley of California. Plant Dis 78:200–207
Nitschke T (1867) Pyrenomycetes germanici. Breslau
Pildain MB, Novas MV, Carmarán CC (2005) Evaluation of anamorphic state, wood decay and production of lignin-modifying enzymes for diatrypaceous fungi from Argentina. J Agric Technol 1:81–96
Pitt WM, Huang R, Trouillas FP, Savocchia S, Steel CC (2010) Evidence that Eutypa lata and other diatrypaceous species occur in New South Wales vineyards. Australas Plant Pathol 39:97–106
Pollock DD, Zwickl DJ, McGuire JA, Hillis DM (2002) Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol 51:664–671
Rannala B, Huelsenbeck JP, Yang Z, Nielsen R (1998) Taxon sampling and the accuracy of large phylogenies. Syst Biol 47:702–710
Rappaz F (1987) Taxonomie et nomenclature des Diatrypacées à asques octosporées. Mycol Helv 2:285–648
Rolshausen PE, Trouillas FP, Gubler WD (2004) Identification of Eutypa lata by PCR-RFLP. Plant Dis 88:925–929
Romero AI, Carmarán CC (2003) First contribution to the study of Cryptosphaeria from Argentina. Fungal Divers 12:161–167
Saccardo PA (1882) Sylloge Fungorum. Vol 1
Saccardo PA (1905) Sylloge Fungorum. Vol 3
Saccardo PA (1926) Sylloge Fungorum. Vol 24
Sinclair WA, Lyon HH (2005) Diseases of trees and shrubs, 2nd edn. Cornell University Press, Ithaca, p 659
Sosnowski MR, Lardner R, Wicks TJ, Scott ES (2007) The influence of grapevine cultivar and isolate of Eutypa lata on wood and foliar symptoms. Plant Dis 91:924–931
Spooner BM (1981) New records and species of British microfungi. Trans Br Mycol Soc 76:265–301
Swofford DL (1999) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b4a. Sinauer Associates, Sunderland
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tiffany LH, Gilman JC (1965) Iowa Ascomycetes IV, Diatrypaceae. Iowa State J Sci 40:121–161
Trouillas FP, Gubler WD (2004) Identification and characterization of Eutypa leptoplaca, a new pathogen of grapevine in Northern California. Mycol Res 108:1195–1204
Trouillas FP, Gubler WD (2010) Pathogenicity of Diatrypaceae species in grapevines in California. Plant Dis 94:867–872
Trouillas FP, Úrbez-Torres JR, Gubler WD (2010a) Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 102:319–336
Trouillas FP, Sosnowski MR, Gubler WD (2010b) Two new species of Diatrypaceae from coastal wattle in Coorong National Park, South Australia. Mycosphere 1:183–188
Úrbez-Torres JR, Adams P, Kama J, Gubler WD (2009) Identification, incidence and pathogenicity of fungal species associated with grapevine dieback in Texas. Am J Enol Vitic 60(4):497–507
Vasilyeva LN, Stephenson SL (2004) Pyrenomycetes of the Great Smoky Mountains National Park. I. Diatrype Fr. (Diatrypaceae). Fungal Divers 17:191–201
Vasilyeva LN, Stephenson SL (2005) Pyrenomycetes of the Great Smoky Mountains National Park. II. Diatrypella (Ces. et De Not.) Nitschke and Cryptovalsa Ces et De Not. (Diatrypaceae). Fungal Divers 19:189–200
Vasilyeva LN, Stephenson SL (2006) Pyrenomycetes of the Great Smoky Mountains National Park. III. Cryptosphaeria Ces. et De Not., Eutypa Tul. et C. Tul., and Eutypella (Nitschke) Sacc. (Diatrypaceae). Fungal Divers 22:243–254
Vasilyeva LN, Stephenson SL (2009) The genus Diatrype (Ascomycota, Diatrypaceae) in Arkansas and Texas (USA). Mycotaxon 107:307–313
Wehmeyer LE (1926) A biologic and phylogenetic study of the stromatic sphaeriales. Am J Bot 13:574–645
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White J (eds) PCR Protocols, a guide to methods and applications. Academic, San Diego, pp 315–322
Yuan ZQ (1996) Fungi and associated tree diseases in Melville Island, Northern Territory, Australia. Aust Syst Bot 9:337–360
Zwickl DJ, Hillis DM (2002) Increased taxon sampling greatly reduces phylogenetic error. Syst Biol 51:588–598
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Trouillas, F.P., Pitt, W.M., Sosnowski, M.R. et al. Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia. Fungal Diversity 49, 203–223 (2011). https://doi.org/10.1007/s13225-011-0094-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-011-0094-0