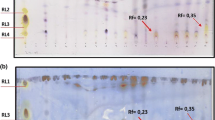
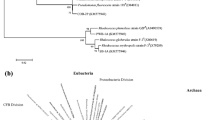
Abstract
Biosurfactant-producing bacteria were isolated from mangrove sediment samples collected in the southern part of Thailand by an enrichment-culture technique in which lubricating oil was the sole carbon source. A total of 1,600 colonies were obtained, which were screened for biosurfactant production using the qualitative drop-collapsing test in a mineral salts medium containing 1% of different carbon sources (commercial sugar, glucose, molasses, and used lubricating oil). Ninety-five isolates were positive for biosurfactant production based on the results of this test, among which 20 could reduce the surface tension of the 48-h culture supernatant. The phylogenetic position of these 20 isolates was evaluated by 16S rRNA gene sequence analysis. The production of biosurfactants was determined for strains representative of eight different bacterial genera. Leucobacter komagatae 183, one of the newly isolated strains showing biosurfactant production, produced extracellular biosurfactants which reduced the surface tension of the culture supernatant from 72.0 to 32.0 m/Nm. Eighteen strains released extracellular emulsifiers able to stabilize the emulsion formed. Among these, the strains L. komagatae 183 and Ochrobactrum anthropi 11/6 exhibited emulsification activities comparable to those of synthetic surfactants. Overall, the new biosurfactant-producing strains isolated in this study display promising features for the future development and use in economically efficient industrial-scale biotechnological processes.

Similar content being viewed by others
References
Abouseoud M, Maachi R, Amrane A, Boudergua S, Nabi A (2008) Evaluation of different carbon and nitrogen sources in production of biosurfactant by Pseudomonas fluorescens. Desalination 223:143–151
Anandaraj B, Thivakaran P (2010) Isolation and production of biosurfactant producing organism from oil spilled soil. J Biosci Tech 1:120–126
Banat IM, Makkar RS, Cameotra SS (2000) Potential commercial applications of microbial surfactants. Appl Microbiol Biotechnol 53:495–508
Batista SB, Mounteer AH, Amorim FR, Totola MR (2006) Isolation and characterization of biosurfactant/bioemulsifier-producing bacteria from petroleum contaminated sites. Bioresource Technol 97:868–875
Bernard D, Pascaline H, Jeremie JJ (1996) Distribution and origin of hydrocarbons in sediments from lagoons with fringing mangrove communities. Mar Pollut Bull 32:734–739
Bicca FC, Fleck LC, Ayub MAZ (1999) Production of biosurfactant by hydrocarbon degrading Rhodococcus ruber and Rhodococcus erythropolis. Rev Microbiol 30:231–236
Blume E, Bischoff M, Reichert JM, Moorman T, Konopka A, Turco RF (2002) Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl Soil Ecol 20:171–181
Bodour AA, Maier RM (2002) Biosurfactants: types, screening methods and application. In: Bitton G (ed) Encyclopedia of environmental microbiology. Wiley, New York, pp 750–769
Bodour AA, Drees KP, Raina MM (2003) Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid Southwestern soils. Appl Environ Microbiol 69:3280–3287
Burgos-Diaz C, Pons R, Espuny MJ, Aranda FJ, Teruel JA, Manresa A, Ortiz A, Marques AM (2011) Isolation and partial characterization of a biosurfactant mixture produced by Sphingobacterium sp. isolated from soil. J Colloid Interf Sci 361:195–204
Burns KA, Garrity SD, Levings SC (1993) How many years until mangrove ecosystems recover from catastrophic oil-spills. Mar Pollut Bull 26:239–248
Chayabutra C, Wu J, Ju LK (2001) Rhamnolipid production by Pseudomonas aeruginosa under denitrification: effects of limiting nutrients and carbon substrates. Biotechnol Bioeng 72:25–33
Cooper DG (1986) Biosurfactants. Microbiol Sci 3:145–149
Cooper DG, Zajic JE, Gerson DF (1979) Production of surface active lipids by Corynebacterium lepus. Appl Environ Microbiol 37:4–10
Darvishi P, Ayatollahi S, Mowla D, Niazi A (2011) Biosurfactant production under extreme environmental conditions by an efficient microbial consortium, ERCPPI-2. Colloid Surface B 84:292–300
Das P, Mukherjee S, Sen R (2009) Substrate dependent production of extracelullar biosurfactant by a marine bacterium. Bioresouce Technol 100:1015–1019
Das P, Mukherjee S, Sivapathasekaran S, Sen R (2010) Microbial surfactants of marine origin: potentials and prospects. Adv Exp Med Biol 672:88–101
Desai JD, Banat IM (1997) Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev 61:47–64
De Acevedo GT, McInerney MJ (1996) Emulsifying activity in thermophilic and extremely thermophilic microorganisms. J Ind Microbiol 16:1–7
From C, Hormazabal V, Hardy SP, Granum PE (2007) Cytotoxicity in Bacillus mojavensis is abolished following loss of surfactin synthesis: Implications for assessment of toxicity and food poisoning potential. Int J Food Microbiol 117:43–49
Gandhimathi R, Kiran GS, Hema TA, Selvin J, Raviji TR, Shanmughapriya S (2009) Production and characterization of lipopeptide biosurfactant by a sponge-associated marine actinomycetes Nocardiopsis alba MSA10. Bioprocess Biosyst Eng 32:825–835
Gudina EJ, Teixeira JA, Rodrigues LR (2010) Isolation and functional characterization of a biosurfactant produced by Lactobacillus paracasei. Colloid Surface B 76:298–304
Huy NQ, Jin S, Amada K, Haruki M, Huu NB, Hang DT, Ha DT, Imanaka T, Morikawa M, Kanaya S (1999) Characterization of petroleum degrading bacteria from oil-contaminated sites in Vietnam. J Biosci Bioeng 88:100–102
Jachimska B, Lunkenheimer K, Malysa K (1995) Effect of position of the functional group on the equilibrium and surface properties of butyl alcohols. J Colloid Interf Sci 176:31–38
Karanth NGK, Deo PG, Veenanadig NK (1999) Microbial production of biosurfactant and their importance. Curr Sci 77:126–166
Ke L, Wang WQ, Wong TW, Wong YS, Tam NF (2003) Removal of pyrene from contaminated sediments by mangrove microcosms. Chemosphere 52:1581–1591
Kebbouche-Gana K, Gana ML, Khemili S, Naimi FF, Bouanane NA, Penninckx M, Hacene H (2009) Isolation and characterization of halophilic archaea able to produce biosurfactants. J Ind Microbiol Biotechnol 36:727–738
Konishi M, Nagahama T, Fukuoka T, Morita T, Imura T, Kitamoto D, Hatada Y (2011) Yeast extract stimulates production of glycolipid biosurfactants, mannosylerythritol lipids, by Pseudozyma hubeiensis SY62. J Biosci Bioeng 111:702–705
Maneerat S (2005) Biosurfactants from marine microorganisms. Songklanakarin J Sci Technol 27:1263–1272
Maneerat S, Phetrong K (2007) Isolation of biosurfactant-producing marine bacteria and characteristics of selected biosurfactant. Songklanakarin J Sci Technol 29:781–791
Maneerat S, Bamba T, Harada K, Kobayashi A, Yamada H, Kawai K (2006) A novel crude oil emulsifier extracted in the culture supernatant of a marine bacterium, Myroides sp. SM7. Appl Microbiol Biotechnol 70:254–259
Mercade ME, Monleon L, de Andres C, Rodon I, Martinez E, Espuny MJ, Manresa A (1996) Screening and selection of surfactant-producing bacteria from waste lubricating oil. J Appl Bacteriol 81:161–166
Nilsson WB, Strom MS (2002) Detection and identification of bacterial pathogens of fish in kidney tissue using terminal restriction length polymorphism (T-RFLP) analysis of 16S rRNA genes. Dis Aquat Org 48:175–185
Olivera NL, Commendatore MG, Delgado O, Esteves JL (2003) Microbial characterization and hydrocarbon biodegradation potential of natural bilge waste microflora. J Ind Microbiol Biotechnol 30:542–548
Pansiripat S, Pornsunthorntaweea O, Rujiravanit R, Kitiyanana B, Somboonthanate P, Chavadej S (2010) Biosurfactant production by Pseudomonas aeruginosa SP4 using sequencing batch reactors: effect of oil-to-glucose ratio. Biochem Eng J 49:185–191
Phalakornkule C, Tanasupawat S (2006) Characterization of lactic acid bacteria from traditional Thai sausages. J Cult Collect 5:46–57
Plaza GA, Zjawiony I, Banat IM (2006) Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon contaminated and bioremediated soils. J Pet Sci Eng 50:71–77
Rahman KSM, Banat IM, Thahira J, Thayumanvan T, Akshmanaperumalsamy P (2002) Bioremediation of gasoline contaminated soil by a bacterial consortium amended with poultry litter, coir pith and rhamnolipid biosurfactant. Bioresource Technol 81:25–32
Rivardo F, Turner RJ, Allegrone G, Ceri H, Martinotti MG (2009) Anti-adhesion activity of two biosurfactants produced by Bacillus spp. prevents biofilm formation of human bacterial pathogens. Appl Microbiol Biotechnol 83:541–553
Rodrigues LR, Teixeira JA, van der Meib HC, Oliveira R (2006) Isolation and partial characterization of a biosurfactant produced by Streptococcus thermophilus A. Colloid Surface B 53:105–112
Rosenberg E, Ron EZ (1998) Surface active polymers from the genus Acinetobacter. In: Kaplan DL (ed) Biopolymers from renewable resources. Springer, Berlin, pp 281–291
Rosenberg E, Ziclerberg A, Rubinowitz C, Gutnick DL (1979) Emulsifier of Arthrobacter RAG-1: isolation and emulsifying properties. Appl Environ Microbiol 37:402–408
Ruggeri C, Franzetti A, Bestetti G, Caredda P, La Colla P, Pintus M, Sergi S, Tamburini E (2009) Isolation and characterization of surface active compound-producing bacteria from hydrocarbon-contaminated environments. Int Biodeter Biodegr 63:936–942
Saimmai A, Sobhon V, Maneerat S (2011) Molasses a whole medium for bosurfactants production by Bacillus strains and their application. Appl Biochem Biotech 165:315–335
Singh BR, Dwivedi S, Al-Khedhairy AA, Musarrat J (2011) Synthesis of stable cadmium sulfide nanoparticles using surfactin produced by Bacillus amyloliquifaciens strain KSU-109. Colloid Surface B 85:207–213
Skorupska A, Janczarek M, Marczak M, Mazur A, Krol J (2006) Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb Cell Fact 16:5–7
Snape I, Ferguson SH, Harvey PM, Riddle MJ (2006) Investigation of evaporation and biodegradation of fuel spills in Antarctica: II Extent of natural attenuation at Casey Station. Chemosphere 63:89–98
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibbons TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTALX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Widada J, Nojiri H, Kasuga K, Yoshida T, Habe H, Omori T (2002) Molecular detection and diversity of polycyclic aromatic hydrocarbon-degrading bacteria isolated from geographically diverse sites. Appl Microbiol Biotechnol 58:202–209
Willumsen PA, Karlson U (1997) Screening of bacteria isolated from PAH-contaminated soils for production of biosurfactants and bioemulsifiers. Biodegradation 7:415–423
Yin B, Gua JD, Wana N (2005) Degradation of indole by enrichment culture and Pseudomonas aeruginosa Gs isolated from mangrove sediment. Int Biodeter Biodegr 56:243–248
Youssef NH, Dunacn KE, Nagle DP, Savage KN, Knapp RM, McInerney MJ (2004) Comparison of methods to detect biosurfactant production by diverse microorganism. J Microbiol Meth 56:339–347
Acknowledgments
The last author would like to thank the Office of the Higher Education Commission, Thailand for financial support for this work through a grant funded under the program Strategic Scholarships for Frontier Research Network for the Ph.D. Program Thai Doctoral degree. This work was also funded by the Faculty of Agro-Industry and Graduate School, Prince of Songkla University, and further supported by the TRF/BIOTEC Special Program for Biodiversity Research and Training grant BRT R651178.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saimmai, A., Tani, A., Sobhon, V. et al. Mangrove sediment, a new source of potential biosurfactant-producing bacteria. Ann Microbiol 62, 1669–1679 (2012). https://doi.org/10.1007/s13213-012-0424-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-012-0424-9