Abstract
Here we show that Bacillus pumilus ICVB403 recently isolated from copepod eggs is able to produce, after 48–72 h of growth in Landy medium, extracellular inhibitory compounds, which are active against Staphylococcus aureus ATCC 25923, methicillin-resistant S. aureus (MRSA) ATCC 43300, MRSA-S1, Staphylococcus epidermidis 11EMB, Staphylococcus warneri 27EMB, and Staphylococcus hominis 13EMB. Moreover, these extracellular inhibitory compound(s) were able to potentiate erythromycin against the aforementioned staphylococci. The minimum inhibitory concentration (MIC) of erythromycin was reduced from 32 μg/mL to 8 μg/mL for MRSA ATCC 43300 and MRSA SA-1 strains, and from 32–64 μg/mL to 4 μg/mL for S. epidermidis 11EMB and S. hominis 13EMB strains.
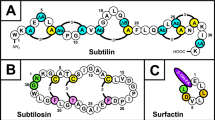
The genome sequencing and analysis of B. pumilus ICVB403 unveiled 3.666.195 nucleotides contained in 22 contigs with a G + C ratio of 42.0%, 3.826 coding sequences, and 73 RNAs. In silico analysis guided identification of two putative genes coding for synthesis of surfactin A, a lipopeptide with 7 amino acids, and for a circular bacteriocin belonging to the circularin A/uberolysin family, respectively.

Similar content being viewed by others
References
Huss HH (2004) Fresh fish quality and quality changes. In: FAO Fisheries Technical Paper No. 348. Food and Agriculture Organization of the United Nations, Rome, Italy, pp 103–109
Food and Agriculture Organization of the United Nations/World Health Organization (FAO/WHO) (2016) La situation mondiale des pêches et aquaculture.
Biji KB, Ravishankar CN, Venkateswarlu R, Mohan CO, Gopal TK (2016) Biogenic amines in seafood: a review. J Food Sci Technol 53(5):2210–2218
Rambla-Alegre M, Miles CO, de la Iglesia P, Fernandez-Tejedor M, Jacobs S, Sioen I, Verbeke W, Samdal IA, Sandvik M, Barbosa V, Tediosi A, Madorran E, Granby K, Kotterman M, Calis T, Diogene J (2018) Occurrence of cyclic imines in European commercial seafood and consumers risk assessment. Environ Res 161:392–398
Davis AR, Rabinson B (2003) Incidence of food borne pathogens on European fish. Food Control 12:67–71
Hosseini H, Misaghi A (2004) Incidence of Vibrio spp. in seafood caught of south coast of Iran. Food Control 8:91–98
Chintagari S, Hazard N, Edwards G, Jadeja R, Janes M (2017) Risks associated with fish and seafood. Microbiol Spectr 5(1). https://doi.org/10.1128/microbiolspec.PFS-0013-2016.
Elbashir S, Parveen S, Schwarz J, Rippen T, Jahncke M, DePaola A (2018) Seafood pathogens and information on antimicrobial resistance: a review. Food Microbiol 70:85–93
Nayak SK (2010) Probiotics and immunity: a fish perspective. Fish Shellfish Immunol 29:2–14
Wang AR, Ran C, Ringo E, Zhou ZG (2017) Progress in fish gastrointestinal microbiota research. Rev Aquacul 2017:626–640. https://doi.org/10.1111/raq.12191
Feldhusen F (2004) The role of seafood in bacterial food borne diseases. Microbes Infect 2:1651–1660
Edwards AM, Massey RC, Clarke SR (2012) Molecular mechanisms of Staphylococcus aureus nasopharyngeal colonization. Mol Oral Microbiol 27:1–10
Atyah MA, Zamri-Saad M, Siti-Zahrah A (2010) First report of methicillin-resistant Staphylococcus aureus from cage-cultured tilapia (Oreochromis niloticus). Vet Microbiol 144:502–504
Soliman MK, Ellakany HF, Gaafar AY, Elbialy AK, Zaki MS,Younes AM (2014) Epidemiology and antimicrobial activity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from Nile tilapia (Oreochromis niloticus) during an outbreak in Egypt. Life Sci J 11(10):1245–1252
Arfatahery N, Mirshafiey A, Abedimohtasab TP, Zeinolabedinizamani M (2015) Study of the prevalence of Staphylococcus aureus in marine and farmed shrimps in Iran aiming the future development of a prophylactic vaccine. Procedia Vaccinol 9:44–49
Pridgeon J, Klesius PH (2012) Major bacterial diseases in aquaculture and their vaccine development. CAB Reviews 2012(048)
Rigos G, Troisi GM (2005) Antibacterial agents in Mediterranean finfish farming: a synopsis of drug pharmacokinetics in important euryhaline fish species and possible environmental implications. Rev Fish Biol Fisher 15(1–2):53–73
Reda RM, Ibrahim RE, Ahmed ENG, El-Bouhy ZM (2013) Effect of oxytetracycline and florfenicol as growth promoters on the health status of cultured Oreochromis niloticus. Egypt J Aquat Res 39(4):241–248
Chanu KV, Thakuria D, Kumar S (2018) Antimicrobial peptides of buffalo and their role in host defenses. Veterinary world 11:192–200
Gomez-Gil B, Roque A, Turnbull JF (2000) The use and selection of probiotic bacteria for use in the culture of larval aquatic organisms. Aquaculture 191:259–270
Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D (2012) Ready for a world without antibiotics? The pensières antibiotic resistance call to action. Antimicrob Resist Infect Control 14:1–11
Drider D, Rebufat S (2011) Prokarytotic antimicrobial peptides: from genes to applications. Springers Editions NY-USA 2011. 451pp
Abriouel H, Franz CM, Omar NB, Gálvez A (2011) Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev 35:201–232
Mondol MAM, Shin HJ, Islam MT (2013) Diversity of secondary metabolites from marine Bacillus species: chemistry and biological activity. Mar Drugs 11(8):2846–2872
Naghmouchi K, Belguesmia Y, Baah J, Teather R, Drider D (2011) Antibacterial activity of class I and IIa bacteriocins combined with polymyxin E against resistant variants of Listeria monocytogenes and Escherichia coli. Res Microbiol 162(2):99–107
Naghmouchi K, Baah J, Hober D, Jouy E, Rubrecht C, Sané F, Drider D (2013) Synergistic effect between colistin and bacteriocins in controlling Gram-negative pathogens and their potential to reduce antibiotic toxicity in mammalian epithelial cells. Antimicrob Agents Chemother 57(6):2719–2725
Zidour M, Chevalier M, Belguesmia Y, Cudennec B, Grard T, Drider D, Souissi S, Flahaut C (2017) Isolation and characterization of bacteria colonizing Acartia tonsa copepod eggs and displaying antagonist effects against Vibrio anguillarum, Vibrio alginolyticus and other pathogenic strains. Front Microbial 8:1919
Drago L, Mattina R, Nicola L, Rodighiero V, De Vecchi E (2011) Macrolide resistance and in vitro selection of resistance to antibiotics in Lactobacillus isolates. J Microbiol 49:651–656. https://doi.org/10.1007/s12275-011-0470-1
The European Committee on Antimicrobial Susceptibility Testing (2018) Breakpoint tables for interpretation of MICs and zone diameters, version 8.0, 2018. http://www.eucast.org/clinical_breakpoints/
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75
Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43(W1):W237–W243
Berkeley R, Heyndrickx M, Logan N, De Vos P (2008) Applications and systematics of Bacillus and relatives. Wiley-Blackwell, Hoboken 133 pp
Liu Y, Lai Q, Dong C, Sun F, Wang L, Li G, Shao Z (2013) Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis. PLoS One 8:e80097
Miranda CA, Martins OB, Clementino MM (2008) Species-level identification of Bacillus strains isolates from marine sediments by conventional biochemical, 16S rRNA gene sequencing and inter-tRNA gene sequence lengths analysis. Antonie Van Leeuwenhoek 93:297–304
Ettoumi B, Raddadi N, Borin S, Daffonchio D, Boudabous A, Cherif A (2009) Diversity and phylogeny of culturable spore-forming bacilli isolated from marine sediments. J Basic Microbiol 49(Suppl 1):S13
Bhate DS (1955) Pumilin, a new antibiotic from Bacillus pumilus. Nature 175(4462):816–817
Brack C, Mikolasch A, Schlueter R, Otto A, Becher D, Wegner U, Albrecht D, Riedel K, Schauer F (2015) Antibacterial metabolites and bacteriolytic enzymes produced by Bacillus pumilus during bacteriolysis of Arthrobacter citreus. Mar Biotechnol (NY) 17:290–304
Kalinovskaya NI, Kuznetsova TA, Ivanova EP, Romanenko LA, Voinov VG, Huth F, Laatsch H (2002) Characterization of surfactin-like cyclic depsipeptides synthesized by Bacillus pumilus from ascidian Halocynthia aurantium. Mar Biotechnol (NY). 4:179–188
Aunpad R, Na-Bangchang K (2007) Pumilicin 4, a novel bacteriocin with anti-MRSA and anti-VRE activity produced by newly isolated bacteria Bacillus pumilus strainWAPB4. Curr Microbiol 55:308–313
Ismail-Ben Ali A, El Bour M, Ktari L, Bolhuis H, Ahmed M, Boudabbous A, Stal LJ (2012) Jania rubens associated bacteria, molecular identification and antimicrobial activity. J Appl Phycol 24:525–534
Olmos J, Paniagua-Michel J (2014) Bacillus subtilis a potential probiotic bacterium to formulate functional feeds for aquaculture. J Microb Biochem Technol 6:361–365
Sreenivasulu P, Suman Joshi DSD, Narendra K, Venkata Rao G, Krishna Satya A (2016) Bacillus pumilus as a potential probiotic for shrimp culture. Int J Fish Aquat Stud 4:107–110
Prieto ML, O’Sullivan L, Tan SP, McLoughlin P, Hughes H, Gutierrez M, Lane JA, Hickey RM, Lawlor PG, Gardiner GE (2014) In vitro assessment of marine Bacillus for use as livestock probiotics. Mar Drugs 12:2422–2445
Landy M, Warren GH, Roseman SB, Golio LG (1948) Bacillomycin, an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc Soc Exp Biol Med 67:539–541
Naruse N, Tenmyo O, Kobaru S, Kamei H, Miyaki T, Konishi M, Oki T (1990) Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity. J Antibiot (Tokyo) 43:267–280
Grangemard I, Peypoux F, Wallach J, Das BC, Labbé H, Caille A, Genest M, Maget-Dana R, Ptak M, Bonmatin JM (1997) Lipopeptides with improved properties: structure by NMR, purification by HPLC and structure-activity relationships of new isoleucyl-rich surfactins. J Pept Sci 3:145–154
Jemil N, Manresa A, Rabanal F, Ben Ayed H, Hmidet N, Nasri M (2017) Structural characterization and identification of cyclic lipopeptides produced by Bacillus methylotrophicus DCS1 strain. J Chromatogr B Analyt Technol Biomed Life Sci 1060:374–386
Arima K, Kakinuma A, Tamura G (1968) Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun 31:488–494
Chen H, Wang L, Su CX, Gong GH, Wang P, Yu ZL (2008) Isolation and characterization of lipopeptide antibiotics produced by Bacillus subtilis. Lett Appl Microbiol 47:180–186
Ndlovu T, Rautenbach M, Vosloo JA, Khan S, Khan W (2017) Characterisation and antimicrobial activity of biosurfactant extracts produced by Bacillus amyloliquefaciens and Pseudomonas aeruginosa isolated from a wastewater treatment plant. AMB Express 7:108
Perez KJ, Viana J, dos S, Lopes FC, Pereira JQ, dos Santos DM, Oliveira JS, Velho RV, Crispim SM, Nicoli JR, Brandelli A, Nardi RMD (2017) Bacillus spp. isolated from Puba as a source of biosurfactants and antimicrobial lipopeptides. Front Microbiol 8:61
Kawai Y, Kemperman R, Kok J, Saito T (2004) The circular bacteriocins gassericin a and circularin a. Curr Protein Pept Sci 5:393–398
Wirawan RE, Swanson KM, Kleffmann T, Jack RW, Tagg JR (2007) Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619–1630
Gálvez A, Maqueda M, Valdivia E, Quesada A, Montoya E (1986) Characterization and partial purification of a broad spectrum antibiotic AS-48 produced by Streptococcus faecalis. Can J Microbiol 32:765–771
Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC (2008) Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl Environ Microbiol 74:4756–4763
Ananou S, Valdivia E, Martínez Bueno M, Gálvez A, Maqueda M (2004) Effect of combined physico-chemical preservatives on enterocin AS-48 activity against the enterotoxigenic Staphylococcus aureus CECT 976 strain. J Appl Microbiol 97:48–56
Arslan S, Özdemir F (2017) Molecular characterization and detection of enterotoxins, methicillin resistance genes and antimicrobial resistance of Staphylococcus aureus from fish and ground beef. Pol J Vet Sci 20:85–94
Rios AC, Moutinho CG, Pinto FC, Del Fiol FS, Jozala A, Chaud MV, Vila MM, Teixeira JA, Balcão VM (2016) Alternatives to overcoming bacterial resistances: state-of-the-art. Microbiol Res 191:51–80
Acknowledgements
We would like to thank Dr. Matthieu Duban and Dr. Gabrielle Chataîgnée for their helpful assistance in the genome analysis and mass spectrometry analysis. We thank past and present members of the group of LOG COPEFISH team (SS) for their involvement in maintaining several cultures of copepods and algae and the Communauté d’Agglomération du Boulonnais (CAB) for supporting the implementation of a copepod-rearing pilot project (agreement Lille University-CAB).
Funding
This work is partly supported by CPER/FEDER Alibiotech grant (2016-2020) from la Région des Hauts-de-France. This work is a contribution to the project CPER 2014-2020 MARCO funded by the French government and the region Hauts-de-France.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All authors of this paper have read and approved the final version submitted. The contents of this manuscript have not been copyrighted or published previously. No procedures performed in these studies have been conducted in human participants and/or animals.
Electronic supplementary material
ESM 1
(DOCX 191 kb)
Rights and permissions
About this article
Cite this article
Zidour, M., Belguesmia, Y., Cudennec, B. et al. Genome Sequencing and Analysis of Bacillus pumilus ICVB403 Isolated from Acartia tonsa Copepod Eggs Revealed Surfactin and Bacteriocin Production: Insights on Anti-Staphylococcus Activity. Probiotics & Antimicro. Prot. 11, 990–998 (2019). https://doi.org/10.1007/s12602-018-9461-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-018-9461-4