Abstract
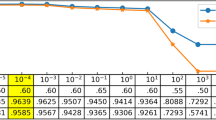
Neuropeptides (NPs) are short secreted peptides produced mainly in the nervous system and digestive system. They activate signaling cascades to control a wide range of biological functions, such as metabolism, sensation, and behavior. NPs are typically produced from a larger NP precursor (NPP) which includes a signal peptide sequence, one or more NP sequences, and other sequences. With the drastic growth of unknown protein sequences generated in the post-genomic age, it is highly desired to develop computational methods for identifying NPP rapidly and efficiently. In this article, we developed a predictor for NPPs based on optimized sequence composition of single amino acid, dipeptide, and tripeptide. Evaluated with independent data set, the predictor showed excellent performance that achieved an accuracy of 88.65% with AUC of 0.95. The corresponding web server was developed, which is freely available at http://i.uestc.edu.cn/neuropeptide/neuropp/home.html. It can help relevant researchers to screen candidate NP precursor, shorten experimental cycle, and reduce costs.





Similar content being viewed by others
References
Brain SD, Cox HM (2006) Neuropeptides and their receptors: innovative science providing novel therapeutic targets. Br J Pharmacol 147(Suppl 1):S202–S211. https://doi.org/10.1038/sj.bjp.0706461
Insel TR, Young LJ (2000) Neuropeptides and the evolution of social behavior. Curr Opinion Neurobiol 10(6):784–789
Hokfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M (2000) Neuropeptides—an overview. Neuropharmacology 39(8):1337–1356
Funkelstein L, Beinfeld M, Minokadeh A, Zadina J, Hook V (2010) Unique biological function of cathepsin L in secretory vesicles for biosynthesis of neuropeptides. Neuropeptides 44(6):457–466. https://doi.org/10.1016/j.npep.2010.08.003
Jekely G (2013) Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci USA 110(21):8702–8707. https://doi.org/10.1073/pnas.1221833110
Rholam M, Brakch N, Germain D, Thomas DY, Fahy C, Boussetta H, Boileau G, Cohen P (1995) Role of amino acid sequences flanking dibasic cleavage sites in precursor proteolytic processing. The importance of the first residue C-terminal of the cleavage site. Eur J Biochem 227(3):707–714
von Eggelkraut-Gottanka R, Beck-Sickinger AG (2004) Biosynthesis of peptide hormones derived from precursor sequences. Curr Med Chem 11(20):2651–2665
von Heijne G (1990) The signal peptide. J Membr Biol 115(3):195–201
Svensson M, Skold K, Svenningsson P, Andren PE (2003) Peptidomics-based discovery of novel neuropeptides. J Proteome Res 2(2):213–219
Che FY, Biswas R, Fricker LD (2005) Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J Mass Spectrom 40(2):227–237. https://doi.org/10.1002/jms.742
Baggerman G, Boonen K, Verleyen P, De Loof A, Schoofs L (2005) Peptidomic analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J Mass Spectrom 40(2):250–260. https://doi.org/10.1002/jms.744
Loewenstein Y, Raimondo D, Redfern OC, Watson J, Frishman D, Linial M, Orengo C, Thornton J, Tramontano A (2009) Protein function annotation by homology-based inference. Genome Biol 10(2):207. https://doi.org/10.1186/gb-2009-10-2-207
Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, Vierstraete E, Rodriguez-Zas SL, Schoofs L, Robinson GE, Sweedler JV (2006) From the genome to the proteome: uncovering peptides in the Apis brain. Science 314(5799):647–649. https://doi.org/10.1126/science.1124128
Liu F, Baggerman G, Schoofs L, Wets G (2006) Uncovering conserved patterns in bioactive peptides in Metazoa. Peptides 27(12):3137–3153. https://doi.org/10.1016/j.peptides.2006.08.021
Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I (2016) UniProtKB/Swiss-Prot, the manually annotated section of the UniProt knowledgebase: how to use the entry view. Methods Mol Biol 1374:23–54. https://doi.org/10.1007/978-1-4939-3167-5_2
Kim Y, Bark S, Hook V, Bandeira N (2011) NeuroPedia: neuropeptide database and spectral library. Bioinformatics 27(19):2772–2773. https://doi.org/10.1093/bioinformatics/btr445
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Tang Q, Nie F, Kang J, Ding H, Zhou P, Huang J (2015) NIEluter: predicting peptides eluted from HLA class I molecules. J Immunol Methods 422:22–27. https://doi.org/10.1016/j.jim.2015.03.021
He B, Kang J, Ru B, Ding H, Zhou P, Huang J (2016) SABinder: a web service for predicting streptavidin-binding peptides. BioMed Res Int 2016:9175143. https://doi.org/10.1155/2016/9175143
Ding C, Yuan LF, Guo SH, Lin H, Chen W (2012) Identification of mycobacterial membrane proteins and their types using over-represented tripeptide compositions. J Proteomics 77:321–328. https://doi.org/10.1016/j.jprot.2012.09.006
Ding H, Feng PM, Chen W, Lin H (2014) Identification of bacteriophage virion proteins by the ANOVA feature selection and analysis. Mol Biosyst 10(8):2229–2235. https://doi.org/10.1039/c4mb00316k
Ru B, t Hoen PA, Nie F, Lin H, Guo FB, Huang J (2014) PhD7Faster: predicting clones propagating faster from the Ph.D.-7 phage display peptide library. J Bioinform Comput Biol 12(1):1450005. https://doi.org/10.1142/S021972001450005X
Lin H, Ding H (2011) Predicting ion channels and their types by the dipeptide mode of pseudo amino acid composition. J Theor Biol 269(1):64–69. https://doi.org/10.1016/j.jtbi.2010.10.019
Lin H, Deng EZ, Ding H, Chen W, Chou KC (2014) iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res 42(21):12961–12972. https://doi.org/10.1093/nar/gku1019
Chen W, Feng PM, Deng EZ, Lin H, Chou KC (2014) iTIS-PseTNC: a sequence-based predictor for identifying translation initiation site in human genes using pseudo trinucleotide composition. Anal Biochem 462:76–83. https://doi.org/10.1016/j.ab.2014.06.022
Ding H, Deng EZ, Yuan LF, Liu L, Lin H, Chen W, Chou KC (2014) iCTX-type: a sequence-based predictor for identifying the types of conotoxins in targeting ion channels. BioMed Res Int 2014:286419. https://doi.org/10.1155/2014/286419
Chen W, Feng P, Ding H, Lin H, Chou KC (2015) iRNA-Methyl: Identifying N(6)-methyladenosine sites using pseudo nucleotide composition. Anal Biochem 490:26–33. https://doi.org/10.1016/j.ab.2015.08.021
Chen W, Feng PM, Lin H, Chou KC (2014) iSS-PseDNC: identifying splicing sites using pseudo dinucleotide composition. BioMed Res Int 2014:623149. https://doi.org/10.1155/2014/623149
Zhu PP, Li WC, Zhong ZJ, Deng EZ, Ding H, Chen W, Lin H (2015) Predicting the subcellular localization of mycobacterial proteins by incorporating the optimal tripeptides into the general form of pseudo amino acid composition. Mol Biosyst 11(2):558–563. https://doi.org/10.1039/c4mb00645c
Tang H, Chen W, Lin H (2016) Identification of immunoglobulins using Chou’s pseudo amino acid composition with feature selection technique. Mol Biosyst 12(4):1269–1275. https://doi.org/10.1039/c5mb00883b
Ding H, Lin H, Chen W, Li ZQ, Guo FB, Huang J, Rao N (2014) Prediction of protein structural classes based on feature selection technique. Interdiscip Sci Comput Life Sci 6(3):235–240. https://doi.org/10.1007/s12539-013-0205-6
Li N, Kang J, Jiang L, He B, Lin H, Huang J (2017) PSBinder: a web service for predicting polystyrene surface-binding peptides. BioMed Res Int 2017:5. https://doi.org/10.1155/2017/5761517
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol (TIST) 2(3):27
Ofer D, Linial M (2014) NeuroPID: a predictor for identifying neuropeptide precursors from metazoan proteomes. Bioinformatics 30(7):931–940. https://doi.org/10.1093/bioinformatics/btt725
Acknowledgements
The authors are grateful to the anonymous reviewers for their valuable suggestions and comments, which have led to the improvement of this paper. This work was supported by the National Natural Science Foundation of China [61571095] and the Fundamental Research Funds for the Central Universities of China [ZYGX2015Z006].
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kang, J., Fang, Y., Yao, P. et al. NeuroPP: A Tool for the Prediction of Neuropeptide Precursors Based on Optimal Sequence Composition. Interdiscip Sci Comput Life Sci 11, 108–114 (2019). https://doi.org/10.1007/s12539-018-0287-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-018-0287-2