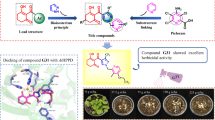
Abstract
Kinetin, a cytokinin which promotes seed germination by inhibiting the action of abscisic acid, is an important molecule known to trigger various molecular mechanisms by interacting with an array of proteins shown from experimental observations in various model organisms. We report here the prediction of most probable protein targets of kinetin from spinach proteome using in silico approaches. Inverse docking and ligand-based similarity search was performed using kinetin as molecule. The former method prioritized six spinach proteins, whereas the latter method provided a list of protein targets retrieved from several model organisms. The most probable protein targets were selected by comparing the rank list of docking and ligand similarity methods. Both of these methods prioritized chitinase as the most probable protein target (ΔG pred = 5.064 kcal/mol) supported by the experimental structure of yeast chitinase 1 complex with kinetin (PDB: 2UY5) and Gliocladium roseum chitinase complex with 3,7-dihydro-1,3,7-trimethyl-1H-purine-2,6-dione (caffeine; 3G6M) which bears a 3D similarity of 0.43 with kinetin. An in vitro study to evaluate the effect of kinetin on spinach seed germination indicated that a very low concentration of kinetin (0.5 mg/l) did not show a significant effect compared to control in inducing seed germination process. Further, higher levels of kinetin (>0.5 mg/l) constituted an antagonist effect on spinach seed germination. It is anticipated that kinetin may have a molecular interaction with prioritized protein targets synthesized during the seed germination process and reduces growth. Thus, it appears that kinetin may not be a suitable hormone for enhancing spinach seed germination in vitro.





Similar content being viewed by others
References
Leubner-Metzger G (2003) Functions and regulation of β-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci Res 13:17–34
Katzman LS, Taylor AG, Langhans RW (2001) Seed enhancements to improve spinach germination. Hortscience 36:979–981
Hartmann HT, Kester DE, Geneve RL, Davies FT (2001) In: Hartmann HT, Kester DE (eds) Hartmann and Kester’s plant propagation: principles and practices. New Jersey, Prentice Hall
Khan AA (1971) Cytokinins: permissive role in seed germination. Science 171:853–859
Al-Khayri JM, Huang FH, Morelock TE (1991) Regeneration of spinach from leaf callus. HortSci 26:913–914
Al-Khayri JM, Huang FH, Morelock TE, Busharar TA (1992) In vitro plant regeneration of spinach from mature seed-derived callus. In Vitro Cell Dev Biol 28:64–66
Robert CE, Douglas GF (1977) Interactions of applied hormones in the germination of Lepidium virginicum seeds. Ohio J Sci 77:236–239
Barciszewski J, Rattan SIS, Siboska G, Clark BFC (1999) Kinetin—45 years on. Plant Sci 148:37–45
Gomez L, Allona I, Casado R, Aragoncillo C (2002) Seed chitinases. Seed Sci Res 12:217–230
Domon JM, Neutelings G, Roger D, David A, David H (2000) A basic chitinase-like protein secreted by embryogenic tissues of Pinus caribaea acts on arabinogalactan proteins extracted from the same cell lines. J Plant Physiol 156:33–39
Krishnaveni S, Liang GH, Muthukrishnan S, Manickam A (1999) Purification and partial characterization of chitinases from sorghum seeds. Plant Sci 144:01–07
Kumar SP, Pandya HA, Jasrai YT (2014) A computational model for non-conserved mature miRNAs from the rice genome. SAR QSAR Environ Res 25:205–220
Kumar SP, Jha PC, Pandya HA, Jasrai YT (2014) Implementation of pseudoreceptor-based pharmacophore queries in the prediction of probable protein targets: explorations in the protein structural profile of Zea mays. Mol BioSyst 10:1833–1844
Kumar SP, Pandya HA, Desai VH, Jasrai YT (2014) Compound prioritization from inverse docking experiment using receptor-centric and ligand-centric methods: a case study on Plasmodium falciparum Fab enzyme. J Mol Recognit 27:215–229
Bernstein FC, Koetzle TF, Williams GJB (1977) The protein data bank: a computer-based archival file for macromolecular structures. J Mol Biol 112:535–542
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Singh UC, Kollman PA (1984) An approach to computing electrostatic charges for molecules. J Comput Chem 5:129–145
Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57:678–683
Bolton E, Wang Y, Thiessen PA, Bryant SH (2008) In: Cornell W (ed) The annual reports in computational chemistry. American Chemical Society, Washington
Wang JC, Chu PY, Chen CM, Lin JH (2012) idTarget: a web server for identifying protein targets of small chemical molecules with robust scoring functions and a divide-and-conquer docking approach. Nucleic Acids Res 40:W393–W399
Chang DT, Oyang YJ, Lin JH (2005) MEDock: a web server for efficient prediction of ligand binding sites based on a novel optimization algorithm. Nucleic Acids Res 33:W233–W238
Wang JC, Lin JH, Chen CM, Perryman AL, Olson AJ (2011) Robust scoring functions for protein–ligand interactions with quantum chemical charge models. J Chem Inf Model 51:2528–2537
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J Am Chem Soc 117:5179–5197
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23:1623–1641
Hospital A, Andrio P, Fenollosa C, Cicin-Sain D, Orozco M, Gelpí JL (2012) MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics 28:1278–1279
Hou X, Du J, Zhang J, Du L, Fang H, Li M (2013) How to improve docking accuracy of AutoDock4.2: a case study using different electrostatic potentials. J Chem Inf Model 53:188–200
Kinnings SL, Jackson RM (2011) ReverseScreen3D: a structure-based ligand matching method to identify protein targets. J Chem Inf Model 51:624–634
Chang CE, Gilson MK (2003) Tork: conformational analysis method for molecules and complexes. J Comput Chem 24:1987–1998
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
The UniProt Consortium (2014) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 42:D191–D198
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99
RAMPAGE Server. Available at: http://ravenbioccam.ac.uk/rampage.php, Accessed May 2014
Lee HS, Zhang Y (2012) BSP-SLIM: a blind low-resolution ligand–protein docking approach using predicted protein structures. Proteins 80:93–110
Anand P, Nagarajan D, Mukherjee S, Chandra N (2014) ABS-Scan: in silico alanine scanning mutagenesis for binding site residues in protein–ligand complex. F1000Research 3:214
Eswar N, Eramian D, Webb B, Shen MY, Sali A (2008) Protein structure modeling with MODELLER. Methods Mol Biol 426:145–159
Shen MY, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152
Jmol: an open-source Java viewer for chemical structures in 3D. Available at http://www.jmol.org/, Accessed March 2015
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Rai S, Sharma M, Jain M, Awasthi A, Purshottam D, Nair N, Sharma A (2010) Rapid in vitro production of cloned plants of Uraria picta (Jacq.) DC—a rare medicinal herb in long-term culture. Appl Biochem Biotechnol 162:1929–1937
Parmar VR, Jasrai YT (2009) Micropropagation of an important medicinal plant Aloe barbadensis Mill (Aloe vera L.) for field plantation. Res J BioTechnol 4:07–10
Chen YZ, Zhi DG (2001) Ligand–protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins 43:217–226
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK (2007) Relating protein pharmacology by ligand chemistry. Nat Biotechnol 25:197–206
Hurtado-Guerrero R, van Aalten DM (2007) Structure of Saccharomyces cerevisiae chitinase 1 and screening-based discovery of potent inhibitors. Chem Biol 14:589–599
Yang J, Gan Z, Lou Z, Tao N, Mi Q, Liang L, Sun Y, Guo Y, Huang X, Zou C, Rao Z, Meng Z, Zhang KQ (2010) Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine. Microbiology 156:3566–3574
Kurepa J, Herouart D, Van Montagu M, Inze D (1997) Differential expression of Cu-, Zn- and Fe-superoxide dismutase genes of tobacco during development, oxidative stress and hormonal treatment. Plant Cell Physiol 38:463–470
Chaitanya KSK, Naithani SC (1998) Kinetin-mediated prolongation of viability in recalcitrant sal (Shorea robusta Gaertn. f.) seeds at low temperature: role of kinetin in delaying membrane deterioration during desiccation-induced injury. J Plant Growth Regul 17:63–69
Theimer RR, Anding G, Matzner P (1976) Kinetin action on the development of microbody enzymes in sunflower cotyledons in the dark. Planta 128:41–47
Xu Y, Gianfagna T, Huang B (2010) Proteomic changes associated with expression of a gene (ipt) controlling cytokinin synthesis for improving heat tolerance in a perennial grass species. J Exp Bot 61:3273–3289
Cavada BS, Moreno FB, da Rocha BA, de Azevedo Jr WF, Castellón RE, Goersch GV, Nagano CS, de Souza EP, Nascimento KS, Radis-Baptista G, Delatorre P, Leroy Y, Toyama MH, Pinto VP, Sampaio AH, Barettino D, Debray H, Calvete JJ, Sanz L (2006) cDNA cloning and 1.75 A crystal structure determination of PPL2, an endochitinase and N-acetylglucosamine-binding hemagglutinin from Parkia platycephala seeds. FEBS J 273:3962–3974
Freeman BC, Beattie GA (2008) An overview of plant defenses against pathogens and herbivores. Plant Health Instr. doi:10.1094/PHI-I-2008-0226-01
Wu CT, Bradford KJ (2003) Class I chitinase and β-1,3-glucanase are differentially regulated by wounding, methyl jasmonate, ethylene, and gibberellin in tomato seeds and leaves. Plant Physiol 133:263–273
Witmer X, Nonogaki H, Beers EP, Bradford KJ, Welbaum GE (2003) Characterization of chitinase activity and gene expression in muskmelon seeds. Seed Sci Res 13:167–178
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8:809–824
van Buuren M, Neuhaus JM, Shinshi H, Ryals J, Meins FJ (1992) The structure and regulation of homeologous tobacco endochitinase genes of Nicotiana sylvestris and N. tomentosiformis origin. Mol Gen Genet 232:460–469
Acknowledgments
Sivakumar Prasanth Kumar acknowledges support from the Department of Science and Technology, New Delhi, India, as INSPIRE Senior Research Fellowship.
Compliance with ethical standards
The authors declare that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sivakumar Prasanth Kumar and Vilas R. Parmar contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 3070 kb)
Rights and permissions
About this article
Cite this article
Kumar, S.P., Parmar, V.R., Jasrai, Y.T. et al. Prediction of protein targets of kinetin using in silico and in vitro methods: a case study on spinach seed germination mechanism. J Chem Biol 8, 95–105 (2015). https://doi.org/10.1007/s12154-015-0135-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12154-015-0135-3