Abstract
Vaccinia-related kinase 1 (VRK1) is one of the mitotic kinases which play key roles in cell cycle control and chromatin modifications. To understand the biological role of the kinase and gain insights into its catalytic mechanism, we performed NMR assignments of catalytically active form of VRK1 with 361 amino acids residues. Here, we present the backbone NMR resonance assignments of the kinase.
Similar content being viewed by others
Biological context
Vaccinia-related kinases (VRKs) are protein kinases that show sequence similarity with vaccinia virus B1 kinase (Nichols and Traktman 2004). Three members of the family have already been identified, of which VRK1 and VRK3 are mainly nuclear kinases while VRK2 is a cytoplasmic protein, found to be associated with the membranes of endoplasmic reticulum. VRK1 protein is widely expressed in different highly proliferative cell lines and also in increased levels different tumor cell lines (Nezu et al. 1997). The kinase plays a major role in regulating cellular stress situations by phosphorylation of several stress related transcription factors, such as p53, c-Jun ATF2 (Santos et al. 2004; Vega et al. 2004; Sevilla et al. 2004; Valbuena et al. 2007). Evidences also support the role of VRK1 protein in the cell cycle entry and progression by phosphorylation of several related proteins such as Rb, PCNA, Cyclin D1 (Kang et al. 2008). Other known targets of the kinase are N-terminal domain of histone H3 and BAF protein, phosphorylation of which are important events in the nuclear condensation event and formation of nuclear envelope during cell division respectively (Gorjanacz et al. 2007; Kang et al. 2007). Recent studies have also identified Coilin, a nuclear protein as a substrate of human VRK1 indicating its role in neurological disorders (Sanz-Garcia et al. 2011). Here, we report the NMR backbone resonance assignments of VRK1.
Methods and experiments
Protein expression and purification
The cDNA fragment encoding human VRK1 protein (1-361) was cloned to pET-29b vector between NdeI and XhoI restriction enzyme sites to generate VRK1 fused with a hexahistidine tag at the C-terminus. Positive clones were checked by DNA sequencing The plasmid DNA was next transformed into E. coli BL21(DE3) expression hosts. The uniformly 15N-labled and 13C/15N-labelled VRK1 proteins were prepared from cells grown in M9 media containing 15N ammonium chloride and [U-13C]-glucose. 2H/13C/15N-labelled VRK1 was prepared from cells grown in the same media with 75 or 100 % D2O. Cells were cultured at 37 °C at 220 rpm until Abs600 of cell culture reached ~0.7 and then induced with 1 mM isopropyl-β-D-thio-galactoside (IPTG) at 25 °C for 4 h. The cell pellet was collected by centrifugation and the protein was purified using Ni2+-NTA affinity column followed by gel filtration chromatography as described previously (Shin et al. 2011).
NMR experiments
NMR samples contained 0.4 mM protein in 20 mM Tris–HCl, pH 6.8, 2 mM DTT, 0.01 % NaN3 and 10 or 100 % D2O. For backbone resonance assignments, transverse relaxation-optimized spectroscopy (TROSY)-based HNCACB, HN(CO)CACB, HNCA, HN(CO)CA, HNCO and HN(CA)CO spectra were obtained using 2H/13C/15N-labeled VRK1 as previously described (Shin et al. 2011). For side chain assignments, TROSY-based 15N-edited NOESY, HCCH-TOCSY and 13C-edited NOESY data were recorded. Selectively labeled NMR samples on 13C/15N-labeled-valine/isoleucine/leucine residues were prepared and HCCH-TOCSY and 13C-edited NOESY experiments were conducted for assigning side chain methyl resonances. NMR experiments were performed on Bruker Avance 800 and Avance 900 MHz spectrometers equipped with cryoprobes at 298 K. All NMR spectra were processed with NMRPipe (Delaglio et al. 1995) and analyzed using SPARKY (T. D. Goddard and D. G. Kneller, UCSF, San Fransico, CA, USA).
NMR assignment and data deposition
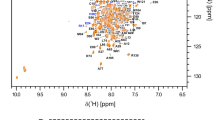
The assignment of the resonance peaks is shown in the Fig. 1. NMR assignments of backbone HN, Cα, Cβ and CO resonances were based on the TROSY-based 3D triple resonance experiments performed on uniformly 2H/13C/15N labeled hVRK1(residues 1-361, including 8 extra C-terminal His-tag sequences, LEHHHHHH). Backbone resonance assignment lacks the 3 amino acid residues (M1, S14, R17) in the N-terminal extension of the protein, which did not appear in the spectrum; besides 4 residues (R358, K359, K360 and E361) in the C-terminal also could not be assigned due to severe overlap. However, the backbone assignment is ~97 % complete of all backbone 1HN, 15N, Cα, Cβ and CO resonances for 345 non-proline residues (335/345 1HN, 335/345 15N, 338/345 Cα, 301/317 Cβ and 338/345 CO). Cα, Cβ and CO resonances of 16 proline residues were also fully assigned by combining TROSY based triple resonance spectra. ~88 % of Hα resonances (317/361) were assigned by the combination of HCCH-TOCSY and 15N-edited NOESY-TROSY using perdeuterated or uniformly 13C/15N labeled hVRK1. Resonances of methyl groups of V, I and L residues were assigned as ~87 % (123/150 of Cγm/Cδm and 138/150 Hγm/Hδm of V, I and L residues, based on the combination of HCCH-TOCSY, 13C-edited NOESY-HSQC, 1H-13C-HMQC, 3D-13C-HMQC-NOESY-HSQC and 15N-edited NOESY-TROSY performed on V/I/L selectively protonated and 2H/13C/15N labeled hVRK1. Prediction of the secondary structures was performed using CSI (Wishart and Sykes 1994), TALOS + program (Shen et al. 2009), and 1HN–1HN NOE patterns based on the 3D-15N-edited NOESY-TROSY spectra. As shown in Fig. 2, secondary structures were estimated by CSI program. The estimated secondary structure of hVRK1 showed typical protein kinase fold similar to other VRK family members (VRK2 and VRK3). The NMR chemical shift data was deposited in the BioMagResBank (http://www.bmrb.wisc.edu, accession number 16738, PDB ID 2KUL). The data may be further used for precise identification and validation of different protein/ligand interactions with the kinase.
References
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Gorjanacz M, Klerkx EP, Galy V, Santarella R, Lopez-Iglesias C, Askjaer P, Mattaj IW (2007) Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J 26(1):132–143. doi:10.1038/sj.emboj.7601470
Kang TH, Park DY, Choi YH, Kim KJ, Yoon HS, Kim KT (2007) Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol 27(24):8533–8546. doi:10.1128/MCB.00018-07
Kang TH, Park DY, Kim W, Kim KT (2008) VRK1 phosphorylates CREB and mediates CCND1 expression. J Cell Sci 121(Pt 18):3035–3041. doi:10.1242/jcs.026757
Nezu J, Oku A, Jones MH, Shimane M (1997) Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 45(2):327–331. doi:S0888754397949387
Nichols RJ, Traktman P (2004) Characterization of three paralogous members of the Mammalian vaccinia related kinase family. J Biol Chem 279(9):7934–7946. doi:10.1074/jbc.M310813200
Santos CR, Vega FM, Blanco S, Barcia R, Lazo PA (2004) The vaccinia virus B1R kinase induces p53 downregulation by an Mdm2-dependent mechanism. Virology 328(2):254–265. doi:10.1016/j.virol.2004.08.013
Sanz-Garcia M, Vazquez-Cedeira M, Kellerman E, Renbaum P, Levy-Lahad E, Lazo PA (2011) Substrate profiling of human vaccinia-related kinases identifies coilin, a Cajal body nuclear protein, as a phosphorylation target with neurological implications. J Proteomics 75(2):548–560. doi:10.1016/j.jprot.2011.08.019
Sevilla A, Santos CR, Vega FM, Lazo PA (2004) Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J Biol Chem 279(26):27458–27465. doi:10.1074/jbc.M401009200
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS + : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44(4):213–223. doi:10.1007/s10858-009-9333-z
Shin J, Chakraborty G, Bharatham N, Kang C, Tochio N, Koshiba S, Kigawa T, Kim W, Kim KT, Yoon HS (2011) NMR solution structure of human vaccinia-related kinase 1 (VRK1) reveals the C-terminal tail essential for its structural stability and autocatalytic activity. J Biol Chem 286(25):22131–22138. doi:10.1074/jbc.M110.200162
Valbuena A, Suarez-Gauthier A, Lopez-Rios F, Lopez-Encuentra A, Blanco S, Fernandez PL, Sanchez-Cespedes M, Lazo PA (2007) Alteration of the VRK1-p53 autoregulatory loop in human lung carcinomas. Lung Cancer 58(3):303–309. doi:10.1016/j.lungcan.2007.06.023
Vega FM, Sevilla A, Lazo PA (2004) p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol Cell Biol 24(23):10366–10380. doi:10.1128/MCB.24.23.10366-10380.2004
Wishart DS, Sykes BD (1994) The 13C chemical shift index. A simple method for the identification of protein secondary structure using 13C chemical shift data. J Biomol NMR 4:171–180
Acknowledgments
This work was generously supported by Ministry of Education of Singapore Academic Research Fund ARC 25/12.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shin, J., Chakraborty, G. & Yoon, H.S. Backbone 1H, 13C and 15N resonance assignments of human vaccinia-related kinase 1 (VRK1). Biomol NMR Assign 8, 29–31 (2014). https://doi.org/10.1007/s12104-012-9446-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-012-9446-2