Abstract
Background and goals
There are no highly sensitive and specific minimally invasive biomarkers for hepatocellular carcinoma (HCC) to date. The objective of this study was to identify serum microRNAs (miRNAs) as potential HCC biomarkers.
Methods
Using miRCURY LNA™ microRNA arrays, the levels of circulating miRNAs in the serum of patients with HCC were compared and controls were matched. Then 253 subjects (112 HCC, 85 chronic hepatitis B [CHB], and 56 healthy controls) were recruited and 12 serum miRNAs were compared by quantitative real-time polymerase chain reaction (qRT-PCR). It was followed by the comparison of serum miRNA concentrations before and after the surgical resection in HCC group.
Results
Median levels of miR-483-5p and miR-500a were higher in HCC patients than in patients with CHB and in healthy controls (p < 0.0001), but there were no differences between CHB patients and healthy controls (p > 0.05) and miR-483-5p levels were significantly reduced in serum samples obtained 30 days after surgical resection (p < 0.0001). The area under receiver operating characteristic curves of miR-483-5p and miR-500a was 74% (cutoff [Ct] value = 2.824, sensitivity = 74%, and specificity = 66%) and 66% (Ct value = 1.830, sensitivity = 74%, and specificity = 51%) for the prediction of HCC, respectively. In detecting HCC, combining α-fetoprotein (AFP) and serum miR-483-5p (sensitivity = 81% and specificity = 83%) was better than AFP alone (sensitivity = 78%, specificity = 70%).
Conclusion
Our observations suggest that serum miR-483-5p and miR-500a might serve as novel, noninvasive biomarkers for HCC. Serum miR-483-5p might complement AFP in detecting HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and the third most common cause of cancer death worldwide [1]. The poor prognosis is partly because there are no reliable markers for early detection. The α-fetoprotein (AFP) is widely used to detect primary HCC but its sensitivity and specificity are disputed [2, 3]. A reliable biomarker to detect HCC at an early stage or with high sensitivity and specificity is needed.
MicroRNAs (miRNAs) are single-stranded, noncoding RNAs of 20–23 nt in length. They play important roles in many cellular processes by bonding to messenger RNAs (mRNAs) and reducing their stability [4]. The miRNAs also act as oncogenes or tumor suppressor genes in many human cancers [5–7] and are key points of accommodation in the development of chronic hepatitis B (CHB) and liver cirrhosis (LC)-related HCC [8–10]. MiRNAs are, thus, reliable biomarkers for such cases of HCC. Tissue-specific miRNAs cannot be used on a wide scale because the procedure is invasive; however, use of serum miRNAs is noninvasive and thus more practical. Serum and other body fluids contain stable miRNA signatures [11, 12], and miRNAs are emerging as potential markers for the diagnosis of several diseases, including cancer [12–17].
To investigate the clinical value of circulating miRNAs in HCC patients, our study first established a detection system and then used the system in 253 subjects. We examined several candidate serum miRNAs, and because CHB is a recognized cause of HCC, we used a group of CHB patients as a control. Our results demonstrate that, in association with AFP, serum miR-483-5p might serve as a dynamic criterion of HCC, excluding the influence of hepatitis B virus (HBV) infection and hepatic cell damage.
Materials and methods
Ethics statement
Written informed consent was obtained from each subject. Independent Ethics Committee approval (ID: 20081006) was obtained from the Ethics Committee of the ZhongShan Hospital, Xiamen University (Xiamen, China).
Study protocol
Our study was performed in five phases: (1) miRCURY LNA™ microRNA array analysis of four individuals (two with HCC and two matched controls); (2) serum miRNA quantification method; (3) determination of serum miRNAs in 112 HCC patients, 85 CHB patients, and 56 healthy controls; (4) data processing and statistical analysis; and (5) comparison of serum miRNA levels before and after the surgical resection of HCC.
Patients, healthy controls, and serum collection
The study has been approved by Zhongshan Hospital, Xiamen University (Xiamen, China). The subjects in the first phase were two HCC patients (one male, one female) and two closely matched healthy controls. In the second phase, serum was obtained from typical HCC, LC, CHB, and colon carcinoma patients matched for age and male-to-female ratio (3:2). Liver-associated clinical indices for all participants are displayed in Table 1 (more details in Table S2). All the individuals involved in this study were confirmed, by abdominal CT, not to have adrenocortical malignance. For serum collection, we used citrate as an anticoagulant and all specimens were stored at –80°C. Sera were collected at Zhongshan Hospital, Xiamen University (Xiamen, China) from 2008 to 2010.
RNA extraction from serum
The RNA was extracted from 400 μL of serum using an Ambion mirVana™ miRNA Isolation Kit (Part Number AM1556), largely according to the protocol provided by the manufacturer. Because serum contains high levels of protein, we mixed 400 μL of serum with an equal volume of 2× Denaturing Solution at room temperature and vortexed it for 10 min to increase the yield of RNA. The final elution solution volume was 50 μL.
miRNA array
The RNA samples were labeled using a miRCURY™ Hy3™/Hy5™ Power Labeling Kit and hybridized on a miRCURY™ LNA Array (Version 14.0). The samples were hybridized on a hybridization station, following the manufacturer’s protocol. Scanning was performed with an Axon GenePix 4000B microarray scanner. GenePix pro Version 6.0 was used to read the raw intensity of the image. The intensity of the green signal was calculated after background subtraction and replicated spots on the same slide were averaged to obtain a median intensity. We used a median normalization method to obtain “normalized data,” where normalized data = ([foreground − background]/median); the median is the 50% quantile of miRNA intensity, which was larger than 50 in all samples after background correction. The miRCURY LNA™ microRNA arrays contain probes for all organisms and viruses listed in miRBase, and have very high miRBase coverage. The capture probes were locked nucleic acid (LNA)-enhanced oligonucleotides. By varying the LNA content and the length of the capture probes, the probes were Tm-normalized for hybridization.
Conversion of RNA to cDNA and qRT-PCR of miRNAs
For each sample, we used 5 μL RNA solution for reverse transcription. Besides the 5 μL RNA template, the 20 μL final volume contained 0.1 μL MMLV (Fermentas), 0.4 μL of 10 μM dNTP mixture (Takara), 4 μL 5× MMLV buffer (Fermentas), 0.1 μL RNAsin (Takara), and 1.6 μL of 125 nM miRNA special stem-loop reverse primer (Invitrogen) (Table S1); water was added to a final volume of 20 μL. The reverse transcriptase conditions were 16°C for 15 min, 55°C for 1 h, and 85°C for 10 min, followed by storage at –80°C until the analysis. Because there are no widely accepted internal references for serum miRNAs, we used a standard curve and artificial reference Ath-miR-156a for absolute quantitation (copy/mL) (for details, see “Results” section). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed using stem-loop special primers (Invitrogen) and TaqMan-based probes (Invitrogen). All oligos used in the study are listed in Table S1. About 20 μL of PCR solution contained 2 μL of reverse transcription product, 2 μL 10× ExTaq buffer (Takara), 0.4 μL of 10 μM dNTP mixture (Takara), 0.4 μL miRNA special TaqMan probe, 0.1 μL ExTaq DNA polymerase, 4 μL Betaine solution (Sigma), 0.8 μL of 5 μM F and R primer mixture, and 10.3 μL water. The qRT-PCR was performed on a BIO-RAD CFX-96™ real-time system at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, and 75°C for 10 s. All reactions were performed in triplicate and Ct values greater than 38 were treated as negative. All miRNA mimics used in this study were purchased from GenePharma, China.
AFP, Hepatitis B surface antigen, and alanine aminotransferase measurements
The AFP, hepatitis B surface antigen, and alanine aminotransferase were measured using commercial kits according to manufacturers’ protocols.
Statistical analysis
The Mann–Whitney test was performed to determine the significance of differences in serum miRNA levels between two groups; for three groups, we used the Kruskal–Wallis test. The receiver operating characteristic (ROC) curves and AUCs were obtained to determine the diagnostic efficacy of serum miRNAs. Multivariate regression analysis was performed. The p values <0.05 were considered statistically significant. All statistical calculations were performed using the SPSS software (Version 11.0).
Results
miRNA array detection of serum miRNAs
The initial aim of our study was to identify HCC-related serum miRNAs and investigate their biomarker potential. Because there is little information on this in the literature, we used miRCURY™ LNA arrays to compare the serum miRNA pool of two representative HCC patients with two matched healthy donors to guide our subsequent research. The heat map, in Fig. 1, shows the result of two-way hierarchical clustering of genes and samples. Each row and column represents a miRNA and a sample, respectively. The miRNA clustering tree is shown on the left and the sample clustering tree on the top. The color scale shown at the bottom illustrates the relative expression levels of miRNAs: red represents a high level; green represents a low level. The normalized data are shown in Table S3.
Correlation of normalized serum miRNA expression shows that the differences between the two HCC patients and the two controls were significant (Fig. S1). However, when we screened the up- and down-regulated miRNAs (threshold value ≥1.5-fold), we found that only miR-483-5p (average fold = +4.00), miRPlus-E1151 (average fold = +2.44), miR-675 (average fold = +1.76), and miR-508-5p (average fold = –2.16) were coincidental among the two HCC patients and the two controls (Figs. 1 and S4 and Table S3).
Serum miRNA quantitation
Although miR-16, U6, and other miRNAs have been used as internal references for serum miRNA in many studies, their stability and origin remain unclear [18]. To prevent the impact of an internal reference on the quantification of serum miRNAs, we used an miRNA mimic standard curve (Figs. 2 and S3) to quantify serum copies normalized to spiked-in Ath-miR-156a [12, 19]. An individual standard preparation was used for each PCR plate reaction. In the statistical analysis, we treated N/A and low (>38) Ct values as half of the lower limit (5 copies/mL).
Increased serum miR-483-5p and miR-500a in HCC patients
To screen serum miRNAs that might serve as specific biomarkers for HCC, we chose miR-483-5p, miR-675, miR-508-5p (our array data), miR-122, miR-223, miR-885-5p, miR-500a, miR-155, miR-21, miR-423-5p, miR-375, and miR-92a (serum miRNAs reported by other authors that may serve as serum biomarkers for cancers [20–26]) as our candidates. We used a different serum cohort, comprising 112 HCC patients of whom 79 were HBV carriers, 56 healthy individuals, and 85 CHB patients. In this phase, all serum miRNA levels were measured in terms of copies/mL according to the standard curve and spiked-in the miRNA mimic.
The qRT-PCR results showed greater serum miR-483-5p and miR-500a in the HCC group than in the CHB group and in the healthy controls, but the differences between the CHB group and the healthy controls were not statistically significant (Fig. 3a, b). On the basis of this finding, we put the CHB group and the healthy control group together as a new non-HCC group. The Mann–Whitney test showed significant differences in serum miR-483-5p and miR-500a between the HCC group and the non-HCC group (both p < 0.0001). However, no such differences were found for other serum miRNAs (data not shown).
Increased serum miR-483-5p and miR-500a in HCC patients (a, b). Serum miR-483-5p (a) and miR-500a (b) levels in 112 HCC patients, 85 CHB patients, and 56 healthy controls determined using qRT-PCR. All levels were quantified using a standard curve and normalized to spiked-in Ath-miR-156a in terms of copies/mL. We treated N/A as half of our lower limit (5 copies/mL). The Mann–Whitney or Kruskal–Wallis test was used to determine statistical significance (*** p < 0.0001, ** p < 0.001, * p < 0.01)
Serum miRNAs as potential auxiliaries in the diagnosis of HCC
To evaluate whether serum miRNAs have diagnostic value, we first performed ROC curve analysis on the data (Fig. 4). This showed that serum miR-483-5p and miR-500a levels had the potential to discriminate HCC patients from non-HCC controls; the areas under the ROC curves were 74% (95% CI = 68–81) and 66% (95% CI = 59–73), respectively, compared with an AUC for AFP of 82% (95% CI = 75–87) (Fig. 4). We excluded serum miR-500a from further analysis because its levels and AUC were low. Serum miR-483-5p had almost the same efficacy as AFP in distinguishing HCC patients from non-HCC patients. More importantly, when we selected 2.824 as a Ct value for serum miR-483-5p (sensitivity = 74% and specificity = 66%, Fig. 4), there were 28 AFP negative (Ct = 20 ng/mL) patients positive for miR-483-5p (Table S2) among a total of 34 AFP-negative HCC patients. We, thus, presumed that we could use miR-483-5p as auxiliary diagnostic index to increase the sensitivity and specificity of AFP. Using the formula (lg[AFP] + lg[miR-483-5p]) as an index for HCC, the AUC was 92% (95% CI = 86–97) (Fig. 4); when the Ct is 3.782, the sensitivity is 81% and the specificity 83%, which is better than that of AFP (79 and 71%, respectively; Table 2). Following that, we performed ROC analysis in both AFP positive (>20 ng/mL) individuals and in AFP negative (<20 ng/mL) individuals; the results further strengthened the conclusion that miR-483-5p has an equal or even greater efficient in diagnosing HCC with negative serum AFP level (Fig. 5). HCC Multivariate regression analysis showed that miR-500a and serum miR-483-5p were correlated to HCC with OR = 1.66 and 4.22 (Table 3).
The ROC curve analysis of miR-483-5p in AFP-positive (>20 ng/mL) HCC patients (a) and in AFP-negative (<20 ng/mL) HCC patients (b). We use AFP = 20 ng/mL as a cutoff to cut all the individuals in three groups into two classes and perform ROC curve analysis of miR-483-5p in them (a for AFP positive and b for AFP negative)
Great decrease of serum miR-483-5p after the surgical resection of HCC
To investigate whether the increased miR-483-5p in HCC patients was due to the development of HCC tumor, we compared miR-483-5p levels before and after the surgical resection in 14 patients with high miR-483-5p level (before the surgical resection). Results showed that miR-483-5p decreased in 100% (14/14) of HCC patients after surgical resection (Fig. 6a). Similar tests were also performed to investigate serum miR-500a but no such trends were seen (data not shown). It is worth noting that, serum miR-483-5p levels increased greatly again before their second surgical operation in two recurrence HCC patients (2 years and 1 year from their first operation each). The AFP profile (Fig. 6b) was also included in the figure as a comparison.
Discussion
The HCC is one of the most malignant cancers. Although there have been many studies in the field of HCC diagnosis, AFP is currently the only relatively reliable serum biomarker in wide use. However, its sensitivity and specificity in detecting HCC are poor [27], and there remains a need to increase the efficacy of diagnosis.
Circulating miRNAs have been investigated as biomarkers for many diseases. For example, Huang et al. [16] found that plasma miR-29a and miR-92a have significant diagnostic value in advanced neoplasia, and Hu et al. [15] concluded that miR-486, miR-30d, miR-1, and miR-499 can serve as independent predictors of overall survival rate in non-small cell lung cancer. Also, several circulating miRNAs that could act as biomarkers of HCC have emerged in recent studies [20–23]. Gui et al. [20] found that serum miR-885-5p was significantly elevated in the sera of patients with liver pathologies, including HCC, and Qu et al. [23] reported that serum levels of miR-16 and miR-199a were significantly lower in HCC than in patients with chronic liver disease and control subjects. Although there are many circulating miRNAs that could be used to detect HCC, none of these distinguishes HCC from other liver diseases, such as CHB and LC [21, 26, 28]. Quantification of circulating miRNA is another problem faced by all the authors [11], as strategies to detect low levels of miRNAs in serum are complicated and there is no widely accepted protocol. In this study, we established a highly sensitive serum miRNA detection system using a mimic standard curve and spiked-in Ath-miRNA in terms of copies/mL and used it to measure serum miRNA levels. Our data demonstrated that stem-loop primer and TaqMan probe-based qRT-PCR can be used to detect serum miRNA even at very low levels. We also found that miR-483-5p and miR-500a were markedly elevated in HCC patients’ serum, according to miRCURY™ LNA miRNA Arrays and qRT-PCR, and could be used to distinguish HCC patients from controls, including patients with CHB. Although statistical analysis showed that the AUCs of miR-483-5p and miR-500a were smaller than that of AFP, serum miR-483-5p can greatly increase the AUC of AFP (92%) according to the formula (lg[AFP] + lg[miR-483-5p]) and with a higher sensitivity (81%) and specificity (83%). In the latter part of our research, we found that miR-483-5p levels would decrease after surgical operation, which can be considered as indirect evidence that tumor accretion causes elevated miRNA levels in serum.
Further research must be conducted before circulating miRNAs can be used clinically to detect disease. In future studies, the following issues must be addressed. First, where do serum miRNAs come from, or how do organs release miRNAs into the blood? Recent studies have observed significant number of miRNAs in various body fluids, including serum [29–31], and the form of the miRNAs in these fluids is also interesting. Second, is serum miR-483-5p commonly elevated in AFP-negative HCC patients, and when does the elevation begin? In other words, could serum miR-483-5p be used as an early stage biomarker for HCC or the biomarker for recidivism of HCC? In the current study, we could not explain the issue exactly because of the limitation of our collections but in subsequent studies it must be attended to. Third, the qRT-PCR strategy must be fast, efficient, and inexpensive. In light of the low concentration of serum miRNAs, huge volumes of serum, which must not be heparinized blood, have to be used in research. This is difficult in clinical practice. Therefore, a more sensitive qRT-PCR system is also needed [32]. Fourth, detecting miRNA in serum is complicated because there are no generally accepted internal references. Internal references that are used to normalize serum miRNA include miR-16, U6, and 5s, but all are disputed. As a diagnostic reagent internal reference is impossible, more practical measures must be deployed. Our quantification method was a significant attempt but was still far from entirely effective. Fifth, we used CHB patients as a comparison because HBV infection is the major etiological factor of HCC in Asian countries, but HCV-infected HCC is very rare. However, HCV infection is a major occurrence of HCC in the US and European countries; however, whether miR-483-5p could be used in HCV-infected HCC patients still remains a question to be investigated. Therefore, its application for HCV- or non-viral HCCs requires further investigation. Finally, there were many other HCC-related miRNAs that require investigation, such as miR-26a, miR-18, and miR-224. However, we did not use them in our current study because of the limitation of collections and the hindrance of the detection system. We believe that when the detection of serum miRNA becomes more mature it must be an interesting topic to follow.
The MiR-483-5p acts as an oncogene co-expressed with Igf2 located in intron 2 of the Igf2 gene, and its target genes are Socs3 and Tnf-α [33]. Ma et al. also found that miR-483-5p was highly expressed in liver, brain, heart, and kidney. This miRNA has also been reported to be upregulated in other malignant tumors [28, 34]. Interestingly, we have found that miR-483-5p levels were higher in tumors in 10 HCC patients than in matched normal tissues, and as a co-expression host gen, IGF2’s mRNA and protein were also upregulated in these tissues with the same trends of miR-483-5p (Fig. S2). It is a suggestive finding for later functional study of miR-483-5p in HCC. Elevated circulating miR-483-5p has been observed in some breast cancer cases, but not universally [35]; this study is consistent with our preliminary screening. In future research, we wish to determine whether miR-483-5p plays a role in the progress of HCC and whether it could serve as a therapeutic target, as has miR-26a [7]. Although miR-483-5p has been reported to be upregulated in various types of cancer, there are currently no other reports of its upregulation in cancer patients. We wish to know whether differences in serum miR-483-5p occur because the complicated portal system in the liver allows cancer cells to release miRNA into the blood more easily than other cells.
In conclusion, according to our data, serum levels of miR-483-5p were obviously elevated in HCC patients, and miR-483-5p can be used as a solitary dynamic criterion or as an auxiliary to AFP in distinguishing HCC from other liver diseases, including CHB and LC. Although our detection system was far from rigorous, our use of standard curves and spikes for miRNA quantification was a valid attempt. We believe that when the source of serum miRNAs has been elucidated and their quantification becomes exact and inexpensive, this novel class of biomarker will be put into wide clinical use.
References
Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin 2011;61:69–90
Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology 2008;48:2047–2063
Zinkin NT, Grall F, Bhaskar K, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res 2008;14:470–477
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297
Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol 2011;223:102–115
Wu J, Liu Z, Shao C, et al. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res 2011;71:349–359
Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137:1005–1017
Zhang X, Liu S, Hu T, et al. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009;50:490–499
Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett 2007;252:157–170
Murakami Y, Toyoda H, Tanaka M, et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One 2011;6:e16081
Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513–10518
Wang GK, Zhu JQ, Zhang JT, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010;31:659–666
Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 2009;106:4402–4407
Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 2010;28:1721–1726
Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118–126
Vasilescu C, Rossi S, Shimizu M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One 2009;4:e7405
Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit Rev Oncol Hematol 2011;80:193–208
Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010;102:1174–1179
Gui J, Tian Y, Wen X, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–193
Xu J, Wu C, Che X, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 2011;50:136–142
Zhang Y, Jia Y, Zheng R, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem 2010;56:1830–1838
Qu KZ, Zhang K, Li H, et al. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 2011;45:355–360
Yamamoto Y, Kosaka N, Tanaka M, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009;14:529–538
Rosenwald S, Gilad S, Benjamin S, et al. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol 2010;23:814–823
Li LM, Hu ZB, Zhou ZX, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 2010;70:9798–9807
Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology 2005;42:1208–1236
Meyer-Rochow GY, Jackson NE, Conaglen JV, et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocr Relat Cancer 2010;17:835–846
Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–1741
Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 2010;38:7248–7259
Zubakov D, Boersma AW, Choi Y, et al. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Leg Med 2010;124:217–226
Roos PH, Jakubowski N. Methods for the discovery of low-abundance biomarkers for urinary bladder cancer in biological fluids. Bioanalysis 2010;2:295–309
Ma N, Wang X, Qiao Y, et al. Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol Cell Endocrinol 2011;333:96–101
Patterson EE, Holloway AK, Weng J, et al. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 2011;117:1630–1639
Zhao H, Shen J, Medico L, et al. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One 2010;5:e13735
Acknowledgments
This study was supported by grants from the Medical Innovation Project of Fujian Province, China (2009-CXB-55) and the Science and Technology Project of Xiamen, China (3502Z20100002).
Conflicts of interest
The authors confirm that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12072_2012_9341_MOESM1_ESM.tif
Fig. S1. Scatter plot analysis of the miRNA Array (A–F). a HCC-1 versus Control-1; b HCC-1 versus Control-2; c HCC-2 versus Control-1; d HCC-2 versus Control-2; e HCC-1 versus HCC-2; and f Control-1 versus Control-2. (TIFF 1888 kb)
12072_2012_9341_MOESM2_ESM.tif
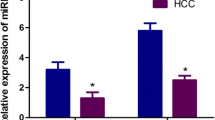
Fig. S2. The miR-483-5p IGF-2 relative expression level in liver tissue. The miR-483-5p relative expression level (according to U6, [a]), IGF-2 expression level (according to GAPDH, [b]), and IGF-2 protein expression level (c) in 10 HCC tissue control to paired normal liver tissue. Primer and probe sequences for real-time PCR: U6, forward: ctgcgcaaggatgaca, reverse: tatggaacgcttcacg. IGF-2, forward: actgcttccaggtgtcatatt, reverse: cctggagacgtactgtgcta. probe: FAM–cggacaacttccccagataccccg-BHQ-1. GAPDH, forward: ggcgatgctggcgctgagtac, reverse: tggtccacacccatgacga. probe: ROX-ttcaccaccatggagaaggctggg-BHQ-2. Antibody used for IGF-2 western blot were purchased from abcam, NO. Anti-IGF2 antibody (ab9574). (TIFF 123 kb)
12072_2012_9341_MOESM3_ESM.tif
Fig. S3. The qRT-PCR amplification curve of miR-483-5p, miR-500a, and Ath-miR-156a (A–C). The qRT-PCR amplification curve of miR-483-5p, miR-500a, and Ath-miR-156a, using miRNAs standard preparation and analysis were performed on CFX Manager (Bio-Rad). Tenfold serial dilution of miRNA was used to generate the standard curves. (TIFF 1958 kb)
Rights and permissions
About this article
Cite this article
Zhang, Z., Ge, S., Wang, X. et al. Serum miR-483-5p as a potential biomarker to detect hepatocellular carcinoma. Hepatol Int 7, 199–207 (2013). https://doi.org/10.1007/s12072-012-9341-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-012-9341-z