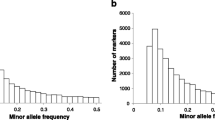
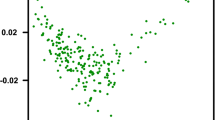
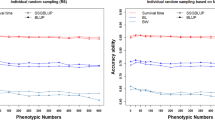
Abstract
A traditional genomewide association study (GWAS) detects genotype–phenotype associations by the vast number of genotyped individuals. This method requires large-scale samples and considerable sequencing costs. Extreme phenotypic sampling proposes make GWAS more cost-efficient and are applied more widely. With extreme phenotypic sampling, we performed a GWAS for n-3 highly unsaturated fatty acids (HUFA) and eviscerated weight (EW) traits in the large yellow croaker population. Of the 32,249 and 29,748 detected SNPs for the two traits, three candidate regions were found in each trait. Three candidate regions associated with HUFA were known near genes on chromosomes 4 and 11, and three candidate regions were on chromosome 6, and 15 for the EW trait. By combing through our GWAS results and the biological functional analysis of the genes, we suggest that the FABP, DGAT, ATP8B1, FAF2 and CERS2 genes, as well as the IGF2, BORA, CYP1A1, GRTP1 and HOX genes are promising candidate genes for n-3 HUFA and EW, respectively, in the large yellow croaker. Moreover, compared with the different numbers of the extreme phenotypic sampling, we conclude that 60% of the extreme phenotypic subsample can obtain a similar result as GWAS with whole phenotypes. Thus, extreme phenotypic sampling could save 40% of the cost for genotyping and DNA extraction without loss of the candidate regions and functional genes. Our study may provide a basis for further genomic breeding and a reference for others who want to perform GWAS with extreme phenotypes.


Similar content being viewed by others
References
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W. et al. 1997 Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Amores A. and Postlethwait J. H. 1998 Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711–1714.
Barnett I. J., Lee S. and Lin X. 2013 Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet. Epidemiol. 37, 142–151.
Bauer R., Voelzmann A., Breiden B., Schepers U., Farwanah H., Hahn I. et al. 2009 Schlank, a member of the ceramide synthase family controls growth and body fat in Drosophila. EMBO J. 28, 3706–3716.
Benjamini Y. and Hochberg Y. 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57, 289–300.
Blanchard P. G., Turcotte V., Côté M., Gélinas Y., Nilsson S., Olivecrona G. et al. 2016 Peroxisome proliferator-activated receptor \(\upgamma \) a ctivation favours selective subcutaneous lipid deposition by coordinately regulating lipoprotein lipase modulators, fatty acid transporters and lipogenic enzymes. Acta Physiol. 217, 227–239.
Brondolin M., Berger S., Reinke M., Tanaka H., Ohshima T., Fuß B. et al. 2013 Identification and expression analysis of the Zebrafish homologs of the ceramide synthase gene family. Dev. Dyn. 242, 189–200.
Browning B. L. and Browning S. R. 2009 A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223.
Chen C., Ai H., Ren J., Li W., Li P., Qiao R. et al. 2011 A global view of porcine transcriptome in three tissues from a full-sib pair with extreme phenotypes in growth and fat deposition by paired-end RNA sequencing. BMC Genomics 12, 448–448.
Cleveland B. M. and Weber G. M. 2016 Effects of steroid treatment on growth, nutrient partitioning, and expression of genes related to growth and nutrient metabolism in adult triploid rainbow trout (Oncorhynchus mykiss). Domest. Anim. Endocrinol. 56, 1–12.
Darias M. J. and Boglino A. 2012 Molecular regulation of both dietary vitamin A and fatty acid absorption and metabolism associated with larval morphogenesis of Senegalese sole (Solea senegalensis). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 161, 130–139.
Dong L., Xiao S., Wang Q. and Wang Z. 2016a Comparative analysis of the GBLUP, emBayesB, and GWAS algorithms to predict genetic values in large yellow croaker (Larimichthys crocea). BMC Genomics 17, 1.
Dong L., Xiao S., Chen J., Wan L. and Wang Z. 2016b Genomic selection using extreme phenotypes and pre-selection of SNPs in large yellow croaker (Larimichthys crocea). Mar. Biotechnol. 18, 575–583.
Duttaroy A. 2009 Transport of fatty acids across the human placenta: a review. Prog. Lipid Res. 48, 52–61.
Feng Y. and Xu Q. 2010 Pivotal role of hmx2, and hmx3, in zebrafish inner ear and lateral line development. Dev. Biol. 339, 507–518.
Fisheries and Fisheries Administration Bureau of the Ministry of Agriculture 2017 China fishery statistical yearbook 2016. China Agricultural Press, Beijing.
Folch J., Lees M. and Sloane-Stanley G. 1957 A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509.
Haas M. J., Shah G. N., Onsteadhaas L. M. and Mooradian A. D. 2014 Identification of ATP8B1 as a blood-brain barrier-enriched protein. Cell Mol. Neurobiol. 34, 473–478.
Hu G., Gu W., Sun P., Bai Q. and Wang B. 2016 Transcriptome analyses reveal lipid metabolic process in liver related to the difference of carcass fat content in rainbow trout (Oncorhynchus mykiss). Int. J. Genomics 2, 1–10.
Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., Daly M. J. et al. 2008 Efficient control of population structure in model organism association mapping. Genetics 178, 1709–1723.
Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., Freimer N. B. et al. 2010 Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354.
Kawanago M., Takemura S., Ishizuka R., Kousaka T. and Shioya I. 2014 Leucine affects growth and hepatic growth-related factor gene expression in Japanese Amberjack. N. Am. J. Aquacult. 76, 415–422.
Lee H. M., Rim H. K., Seo J. H., Kook Y. B., Kim S. K., Oh C. H. et al. 2014 HOX-7 suppresses body weight gain and adipogenesis-related gene expression in high-fat-diet-induced obese mice. BMC Complement. Altern. Med. 14, 1–7.
Li D., Lewinger J. P., Gauderman W. J., Murcray C. E. and Conti D. 2011 Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet. Epidemiol. 35, 790–799.
Li H. and Durbin R. 2010 Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760.
Mohd-Yusof N. Y., Monroig O., Mohd-Adnan A., Wan K. L. and Tocher D. R. 2010 Investigation of highly unsaturated fatty acid metabolism in the Asian sea bass, Lates calcarifer. Fish Physiol. Biochem. 36, 827–843.
Mullen T. D., Hannun Y. A. and Obeid L. M. 2012 Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 441, 789–802.
Murillo E., Rao K. S. and Durant A. A. 2013 The lipid content and fatty acid composition of four eastern central Pacific native fish species. J. Food Compos. Anal. 33, 1–5.
Omar D. L. C., Wen X., Ke B., Song M. and Nicolae D. L. 2009 Gene, region and pathway level analyses in whole-genome studies. Genet. Epidemiol. 34, 222–231.
Pruitt K. and Tatusova T. D. 2005 NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, 501–504.
Ren H. T., Zhang G. Q., Li J. L., Tang Y. K., Li H. X., Yu J. H. et al. 2013 Two \(\Delta \)6-desaturase-like genes in common carp (Cyprinus carpio var. Jian): Structure characterization, mRNA expression, temperature and nutritional regulation. Gene 525, 11–17.
Segura V., Vilhjálmsson B. J., Platt A., Korte A., Seren Ü., Long Q. et al. 2012 An efficient multi-locus mixed model approach for genome-wide association studies in structured populations. Nat. Genet. 44, 825–830.
Shi J. and Walker M. G. 2007 Gene set enrichment analysis (GSEA) for interpreting gene expression profiles. Curr. Bioinform. 2, 133–137.
Sram R. J., Binkova B., Dejmek J., Chvatalova I., Solansky I., Topinka J. et al. 2006 Association of DNA adducts and genotypes with birth weight. Mutat. Res. 608, 121–128.
Thomas Y., Cirillo L., Panbianco C., Martino L., Tavernier N., Schwager F. et al. 2016 Cdk1 phosphorylates SPAT-1/Bora to promote Plk1 activation in C. elegans and Human cells. Cell Rep. 15, 510–518.
Xiao S., Wang P., Dong L., Zhang Y., Han Z., Wang Q. and Wang Z. 2016 Whole-genome single-nucleotide polymorphism (SNP) marker discovery and association analysis with the eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) content by genotyping-by-sequencing (GBS) in teleost Larimichthys crocea. Peerj 4, e2664.
Yamamoto T., Yamaguchi H., Miki H., Kitamura S., Nakada Y., Aicher T. D. et al. 2011 A novel coenzyme A:diacylglycerol acyltransferase 1 inhibitor stimulates lipid metabolism in muscle and lowers weight in animal models of obesity. Eur. J. Pharmacol. 650, 663–672.
Yang J., Jiang H., Yeh C. T., Yu J., Jeddeloh J. A., Dan N. et al. 2015 Extreme-phenotype genome-wide association study (XP-GWAS): a method for identifying trait-associated variants by sequencing pools of individuals selected from a diversity panel. Plant. J. 84, 222–226.
Zhang X., Huang S., Zou F. and Wang W. 2010 TEAM: efficient two-locus epistasis tests in human genome-wide association study. Bioinformatics 26, 217–227.
Zhou Y. J., Wang Y. and Chen L. L. 2016 Detecting the common and individual effects of rare variants on quantitative traits by using extreme phenotype sampling. Genes 7, 1–12.
Zou S. and Jiang X. 2008 Gene duplication and functional evolution of Hox genes in fishes. J. Fish Biol. 73, 329–354.
Acknowledgements
We thank Kun Ye, Shuangbin Xu, Yuxue Gao and other colleagues in the laboratory who participated in fish sampling and measuring the traits. This work was supported by the Key Projects of the Xiamen Southern Ocean Research Centre (14GZY70NF34) and China Agriculture Research System (CARS-47-G04).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Silvia Garagna
Liang Wan and Linsong Dong contributed equally to this work. XW revised the manuscript. ZW conceived the research and revised the manuscript. LW drafted the manuscript. LD performed the analysis and co-authored the manuscript. SX screened SNP loci. ZH analysed gene function. Two reviewers revised our manuscript and gave very valuable advice. All authors read and approved the final manuscript.
Rights and permissions
About this article
Cite this article
Wan, L., Dong, L., Xiao, S. et al. Genomewide association study for economic traits in the large yellow croaker with different numbers of extreme phenotypes. J Genet 97, 887–895 (2018). https://doi.org/10.1007/s12041-018-0973-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-0973-1