Abstract
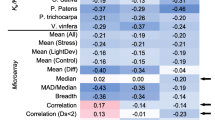
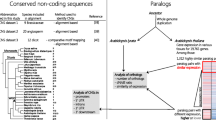
As an index of functional divergence, expression divergence between duplicate gene copies has been observed and correlated with protein coding sequence divergence and bias in gene functional classes. However, the changes in the cis-regulatory region of the duplicate genes which is thought to have important role in expression divergence, has not been explored on the genome-wide scale. We analyzed functional genomics data for a large number of duplicated gene pairs formed by ancient polyploidy events in Arabidopsis thaliana. The divergence in cis-regulatory regions between two copies is positively correlated with the magnitude difference of expression. Moreover, we find that highly expressed duplicate gene pairs have a more diverged cis-regulatory region than weakly expressed gene pairs. We also show that the correlation between expression functional constraint and protein functional constraint is different in old and young duplicate pairs. Our results suggest that cis-regulatory sequence divergence contributes to the expression divergence of duplicate genes formed by genome-wide duplication. Cis-regulatory region diverges faster in highly expressed duplicate pairs. The diversify selection strengths that act on cis-regulatory region and protein coding region are negatively correlated in young duplicate pairs under expression constraint.
Similar content being viewed by others
References
Ohno S. Evolution by Gene Duplication. Berlin, New York: Springer-Verlag, 1970. 160
Zhang J Z. Evolution by gene duplication: An update. Trends Ecol Evol, 2003, 18: 292–298
Lockton S, Gaut B S. Plant conserved non-coding sequences and paralogue evolution. Trends Genet, 2005, 21: 60–65
Blanc G, Wolfe K H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell, 2004, 16: 1679–1691
Reinisch A J, Dong J M, Brubaker C L, et al. A detailed RFLP map of cotton, Gossypium hirsutum × Gossypium barbadense: Chromosome organization and evolution in a disomic polyploid genome. Genetics, 1994, 138: 829–847
Vision T J, Brown D G, Tanksley S D. The origins of genomic duplications in Arabidopsis. Science, 2000, 290: 2114–2117
Simillion C, Vandepoele K, Van Montagu M C, et al. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA, 2002, 99: 13627–13632
Bowers J E, Chapman B A, Rong J, et al. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature, 2003, 422: 433–438
Blanc G, Hokamp K, Wolfe K H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res, 2003, 13: 137–144
Van De Peer Y, Taylor J S, Braasch I, et al. The ghost of selection past: Rates of evolution and functional divergence of anciently duplicated genes. J Mol Evol, 2001, 53: 436–446
Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol, 2002, 19: 1760–1768
Haberer G, Hindemitt T, Meyers B C, et al. Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol, 2004, 136: 3009–3022
Park C, Makova K D. Coding region structural heterogeneity and turnover of transcription start sites contribute to divergence in expression between duplicate genes. Genome Biol, 2009, 10: R10
Lynch M, Conery J S. The evolutionary fate and consequences of duplicate genes. Science, 2000, 290: 1151–1155
Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet, 2004, 20: 544–549.
Casneuf T, De Bodt S, Raes J, et al. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol, 2006, 7: R13
Ganko E W, Meyers B C, Vision T J. Divergence in expression between duplicated genes in Arabidopsis. Mol Bio Evol, 2007, 24: 2298–2309
Schmid M, Davison T S, Henz S R, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet, 2005, 37: 501–506
Simpson P, Ayyar S. Evolution of cis-regulatory sequences in Drosophila. Adv Genet, 2008, 61: 67–106
Castillo-Davis C I, Hartl D L, Achaz G. Cis-regulatory and protein evolution in orthologous and duplicate genes. Genome Res, 2004, 14: 1530–1536
Hurles M. Gene duplication: The genomic trade in spare parts. PLoS Biol, 2004, 2: E206
Gu Z, Cavalcanti A, Chen F C, et al. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol, 2002, 19: 256–262
Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 1994, 22: 4673–4680
Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol, 1994, 11: 725–736
Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci, 1997, 13: 555–556
Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet, 2004, 20: 544–549
Ganko E W, Meyers B C, Vision T J. Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol, 2007, 24: 2298–2309
Pal C, Papp B, Hurst L D. Highly expressed genes in yeast evolve slowly. Genetics, 2001, 158: 927–931
Force A, Lynch M, Pickett F B, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics, 1999, 151: 1531–1545
Blanc G, Wolfe K H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell, 2004, 16: 1679–1691
Casneuf T, De Bodt S, Raes J, et al. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol, 2006, 7: R13
Maere S, De Bodt S, Raes J, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA, 2005, 102: 5454–5459
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Chen, K., Zhang, Y., Tang, T. et al. Cis-regulatory change and expression divergence between duplicate genes formed by genome duplication of Arabidopsis thaliana. Chin. Sci. Bull. 55, 2359–2365 (2010). https://doi.org/10.1007/s11434-010-3027-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-010-3027-5