Abstract
In this article we present the activities of the Ontology Working Group (OWG) under the Metabolomics Standards Initiative (MSI) umbrella. Our endeavour aims to synergise the work of several communities, where independent activities are underway to develop terminologies and databases for metabolomics investigations. We have joined forces to rise to the challenges associated with interpreting and integrating experimental process and data across disparate sources (software and databases, private and public). Our focus is to support the activities of the other MSI working groups by developing a common semantic framework to enable metabolomics-user communities to consistently annotate the experimental process and to enable meaningful exchange of datasets. Our work is accessible via a public webpage and a draft ontology has been posted under the Open Biological Ontology umbrella. At the very outset, we have agreed to minimize duplications across omics domains through extensive liaisons with other communities under the OBO Foundry. This is work in progress and we welcome new participants willing to volunteer their time and expertise to this open effort.
Similar content being viewed by others
Introduction
The storage, management, exchange and description of ‘omics based investigations, such as metabolomics, present challenges to biologists and bioinformaticians (Brooksbank and Quackenbush 2006; Fiehn et al. 2006; Sansone et al. 2006; Shulaev 2006). The Metabolomics Standards Initiative (MSI, http://msi-workgroups.sf.net) Working Groups have recognized that the establishment of reporting standards, such as minimal information requirements and exchange formats with defined semantics are necessary to enable efficient data sharing and meaningful data mining (Castle et al. 2006). Often these requirements are captured as free text, which is subject to ambiguities, redundancy, and typographical errors and as such reduces the power of computational approaches to retrieve the information and unambiguously interpret the experimental procedures.
Adding an interpretive annotation layer to the textual information is commonly done with representational artefacts (RA) such as structured controlled vocabularies (CVs) and/or ontologies (Cimino and Zhu 2006; Schulze-Kremer 1998), consisting of related representational units (RU). A CV is a set of terms (or RU), defined by an authority or through community agreement, in most cases formalized as is-a hierarchy of terms (taxonomy, although within the bio community this term is often used in the more restricted sense of a biological species taxonomy). Each RU is described by means of attributes such as identifier, name, definition and definition source (Smith et al. 2006). A CV is a simple and intuitive way of inserting an interpretive layer of semantics amongst terms used by different experimentalists to describe (annotate) an experimental parameter, in an unambiguous way, for example a type of sample treatment or instrument. Compared to ontologies, CVs are rather informal and lightweight representation artefacts. An ontology is a more explicit and formal representation of the knowledge in a domain, lying at the top end of the semantic complexity scale. Ontologies are semantically rich representations, containing CVs terms as classes as well as their properties, and logical statements for characterising those classes and the ways in which they can or cannot be related to each other.
By way of defined semantics, ontologies provide regimentations of a given terminology, while the defined syntax increases the interoperability between systems exchanging information. Ontologies facilitate the development of systems for data annotation and natural language processing and thereby ontology-based knowledge representations can extend the power of computational approaches to information retrieval, interpretation of experimental procedures, data exploration and knowledge discovery (Blake and Bult 2006). This potential has encouraged several scientific communities, including those operating in the metabolomics domain, to develop ontologies to be used for data annotation (Bodenreider and Stevens 2006; Field and Sansone 2006; Lan et al. 2003; Rubin et al. 2006; Schulze-Kremer 2002; Shulaev 2006; Stevens et al. 2006).
This article describes the working strategy, the developmental phases, the current activities and the challenges of the MSI Ontology Working Group (MSI OWG, http://msi-ontology.sf.net) in its effort to reach a broad consensus in the community on the formal semantic representation that is required to describe metabolomics investigations unambiguously.
The MSI OWG working strategy
The MSI OWG brings together members from diverse backgrounds and perspectives, including metabolomics practitioners, chemometrician, computer scientists, bioinformaticians (data managers, systems developers and data analysts) and ontology engineers.
Scope
The scope of the MSI OWG is to support the activities of the (1) Biological Context Metadata sub-WGs as well as the (2) Chemical Analysis, (3) Data Processing and (4) Exchange Format WGs (Sumner et al. 2007; Morrison et al. 2007; Griffin et al. 2007; van der Werf et al. 2007; Fiehn et al. 2007). The minimal reporting requirements identified by the first three WGs will inform the development of data exchange standards (Hardy and Taylor 2007) in order to provide a common mode of transport for the information between systems. Our work will ultimately provide a formal semantic interpretation for the format, by developing a common semantic framework to enable the metabolomics user communities to consistently annotate the experimental process and ensure meaningful exchange of their datasets. The MSI OWG has been conceived as a ‘single point of focus’ for communities where independent activities—to develop terminologies and databases for metabolomics investigations—are underway. Interoperability among these systems is the key driving force behind this endeavour.
Operating plan
The MSI OWG plans to (1) reach a consensus on a core set of CVs and (2) develop a corresponding ontology. Specifically the CVs and ontology aim to
-
Assist with the representation of study designs, protocols and instrumentation used, data generated and the types of analyses performed on them;
-
Provide a consensual set of terms for the consistent semantic description of data across disparate metabolomics resources (software and databases, both private and public).
The development of the CVs and an ontology for metabolomics is a long iterative process relying on the following stakeholders to provide input:
-
MSI OWG members as developers of the CVs and ontology;
-
Ontology experts/knowledge engineers to provide advice about the engineering of the ontology and practical use cases for an ontology-driven application;
-
Last but not least metabolomics practitioners/domain experts to provide use cases for the ontology, validate the CVs and ontology produced and advise on additional terms to be included.
Operating principles
The MSI OWG operates under the assumption that no one group or community alone can bridge the ‘semantic gap’, and that a synergistic effort is the only way forward. We work cooperatively and maintain a public website with the names of participating members to remain approachable, inclusive and transparent while the size of the group and the complexity of the tasks increase. We communicate via two mailing lists. The first list is open to the public, or those only interested to be kept informed with the progress, while the other list is ‘closed’ and available only to those willing to (1) share the terminology they currently use and (2) invest time and expertise in such collaborative endeavour. Our documents are publicly available via the MSI OWG webpage and drafts of the ontology are posted under the Open Biomedical Ontology (OBO, http://obo.sf.net; Rubin et al. 2006) umbrella. Readers, potential users and developers wishing to send feedback to this and other MSI WGs, can also use the following email address: msi-workgroups-feedback[at]lists.sourceforge.net.
Fortunately, there is a generally accepted view that concerted efforts are required across the functional genomics and system biology communities to work towards harmonised and interoperable reporting standards. At the very outset, we strived to reduce the duplication of efforts across ‘omics domains, where commonality exists, through extensive liaisons with other standards initiatives (described in the next sections) and other ontology communities under the OBO Foundry (Smith et al. under review; http://obofoundry.org). Common standards will benefit the entire scientific community by simplifying the task of data integration and facilitate the work of software developers, vendors and equipment manufacturers by reducing the time involved in and costs of implementing standards-compliant products (Quackenbush 2004).
Developmental phases
Phase 1—Use cases and CV
As described in the section above, ultimately our work will provide a semantic framework for the exchange format, to be agreed upon by the Data Exchange WG, that describes the minimal reporting requirements relevant for the interpretation of metabolomics investigations. Since both the definition of minimal reporting requirements and the development of a data exchange format represent work in progress, our work should be considered explorative and at a very early stage.
Domain coverage and resources
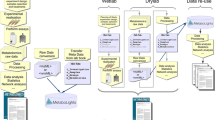
To prioritise our work, we have divided the domain coverage into two main components. Figure 1 shows the components in the investigation workflow: general components (investigation design, origin of the sample and characteristics, sample treatments, sample collection and computational analysis) and the technology-dependant components (instrument-specific sample preparation, instrumental analysis and data pre-processing). Conforming to a generally accepted view that duplication and incompatibility should be avoided, the development of CVs (and a subsequent ontology) for the general investigation components are built as a collaborative effort with standardization initiatives in other ‘omics domains, such as the Human Proteome Organization Proteomics Standards Initiatives (HUPO-PSI, http://psidev.sf.net) (Hermjakob 2006; Taylor et al. 2006) and the Microarray Gene Expression Data Society (MGED, http://www.mged.org) as part of the Ontology for Biomedical Investigations (OBI), further described below. OBI promises to be particularly useful for describing the biological sample and ultimately fulfils the ontological requirements of the MSI Biological Context Metadata sub-WGs. The CV for the technology-dependant components will be our primary focus, starting with the Nuclear Magnetic Resonance (NMR) spectroscopy sub-component. For the Mass Spectrometry (MS) sub-component the OWG will leverage on previous work by the PSI MS Ontology WG. The CVs for the Chromatography sub-component, shared by both proteomics and metabolomics domains, will be developed in close collaboration with the PSI Sample Processing Ontology WG. Every effort will be made to cover as many components as possible and similarly to evaluate and leverage on existing public sources of terms (Allen et al. 1995; Bodenreider 2004; de Matos et al. 2006; Jenkins et al. 2004; Kanehisa et al. 2006; Lindon et al. 2005; Soldatova and King 2006; Spasic et al. 2006; Vranken et al. 2005; Wishart et al. 2007).
Naming conventions and metadata recommendations
At present, neither unified naming conventions, nor common metadata elements have been agreed upon by the ontology-oriented communities for naming and annotating RUs within RAs as well as the RA as a whole (Rickard et al. 2004; Supekar and Musen 2005). Naming conventions prescribe how CV terms and ontological classes should be named and formulated in a consistent manner to unify term appearance, reducing redundancy and increasing precision. These conventions would also provide guidance the ontology engineer on how to handle content related issues, for example Definition and Synonym (semantic naming conventions) and how to tackle lexical issues, such as term/class name length, allowed character set and format, word separators and word tense (syntactic naming conventions). Metadata elements belong to different categories. For example descriptive metadata helps to add useful information on RUs, e.g., definitions or provides examples, while administrative metadata provides information such as when and how a RU or RA was created (release date, version, authority etc.). In the absence of naming conventions and metadata elements applicable to our case, we have started working on such common recommendations in collaboration with the PSI Ontology WGs (Schober et al. 2007). The use of such common conventions would be pivotal in the development and maintenance of the ontology resource by the large participating communities. First drafts of the naming conventions and metadata ontologies are available from our webpage (http://msi-ontology.sourceforge.net/recommendations).
CVs master list
CV master lists for each sub-component will be created iteratively, requiring continuous interactions among the ontology engineers, the domain experts and the other MSI WGs, especially while both the minimal reporting requirements and the format are work in progress. We work according to the following steps:
-
1.
Start from an initial list of terms for a sub-component from a certain resource (database models, glossaries etc.); add definitions for each term and make these compliant to the naming conventions. Keep track of the relationships between the terms (is_a, part_of etc) if provided for the ontology development phase;
-
2.
Structure the terms in an is_a hierarchy (taxonomy) for sorting and redundancy removal;
-
3.
Discuss the CVs within the OWG, and then circulate to the practitioners in the relevant metabolomics area. This will ensure that the lists are as complete as possible, that we obtain valid definitions and will aid ontology construction later on;
-
4.
Explore the use of text mining over a relevant collection of metabolomics papers to identify frequently used terms and enrich the term list;
-
5.
Once general agreement has been reached on the initial CVs, further resources will be processed in turn by deciding which of their terms should be incorporated into the initial CV. For each of these terms synonyms, definitions and relationships will be identified as before;
-
6.
When all resources for a given sub-component have been exhausted, it will be determined which domains remain to be covered. At this stage, we will need to actively collaborate with both the metabolomics practitioners and the other MSI WGs, particularly with the Data Exchange WG, to ensure the quality and completeness of the proposed CV.
Iterative building of such informal ontology models helps to expand our list of terms, relations, their definition or meaning, and additional information such as examples to clarify meaning where appropriate.
Phase 2—Ontology
The OWG’s ultimate goal is to combine the CVs master lists and add further formal structure to create a formal ontology. To achieve this goal, the OWG engages with leading experts in the field of ontology and other ontology communities under the OBO and the OBO Foundry umbrellas. The OBO Foundry is a recent initiative that aims to establish a framework for semantic interoperability in the field of life science. To ensure consistent evolution of the ontologies, the OBO Foundry leaders have issued a set of development recommendations, which will be enhanced in the course of time as new aspects of ontology best practice become established. These recommendations will include the use of (1) an upper formal ontology, OBO Upper Biomedical Ontology (UBO) currently being developed and based on the Basic Formal Ontology (BFO, http://www.ifomis.uni-saarland.de/bfo) to define the top-level class framework under which knowledge representation will be carried out and (2) Relation Ontology (Smith et al. 2005) providing well characterized relations to describe how entities relate to each other (e.g., foundational relations is_a or part_of, but also temporal and spatial relations such as develops_from and located_in). The OBO Foundry also addresses housekeeping needs for ontology maintenance and editing, recommending, among other things Ontology Web Language (OWL, http://www.w3.org/2003/08/owlfaq) and OBO as the format for distribution.
The OWG directly participates in OBI (previously titled FuGO, http://obi.sourceforge.net, Whetzel et al. 2006), an international collaborative project, initiated in 2005, which aims to build a cross-domain ontology as a resource for the annotation of biological, medical and environmental investigations. OBI is an OBO Foundry project set to provide terms that can be used to annotate investigations and the protocols, instrumentation and materials used in those investigations, along with the data generated and analyses performed. OBI brings together HUPO-PSI, MGED Society and other communities, and where the MSI OWG represents the metabolomics domain in this collaborative effort. According to the OBI working strategy, the general experimental components are built collaboratively, while each participating community proposes an informal ontology model relevant to their specific domains. The MSI OWG will provide technology-dependant components by using the relevant OBI “leaf nodes” (e.g., Instrument) as top-level classes. These are then harmonized and positioned within the common BFO top level ontology to ensure reuse and integration with other existing bio-ontologies as described in Rosse et al. 2005. The OBI project is driven by a coordinating committee, bringing together representatives of the participating communities, while guidance on design and engineering is provided by an Advisory Board, including recognised ontology experts. OBI is being developed in OWL using the Protégé ontology editor (Noy et al. 2003). Use cases and terms from each community, minutes of the teleconferences, reports from face-to-face workshops and presentations are available from the project website. An initial version of the top-level structure of OBI, using the BFO is available at the OBO website (http://obo.sourceforge.net/cgi-bin/detail.cgi?obi). A first draft of OBI will be considered ‘completed’ when the general (common) experimental component and all the technology-dependant components have been developed and harmonized (redundancy removed).
Current activities
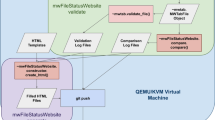
The MSI OWG posts and maintains the ontological components built under the OBO umbrella, in anticipation of OBI being completed. In these first months of our activity, we have focused on NMR experiments in the context of metabolomics investigations. The NMR.owl (available at the OBO site: http://obo.sourceforge.net/cgi-bin/detail.cgi?nmr) is a pure taxonomy of 247 classes in OWL format annotated with metadata through annotation properties. This ontological component encompasses terms of different types: (1) methods, (2) instruments, (3) parameters that can be measured, and other terms. Once collected, the initial terms have been normalised according to the proposed naming conventions (synonyms, acronyms and abbreviations added where known) and taxonomized using the Protégé editor (Fig. 2). Subsequently these have been placed (binned) under the relevant OBI and BFO classes. To populate the initial list of terms and then to refine the ontology, we are currently exploring a text-mining approach over a relevant domain specific collection of MEDLINE abstracts (http://www.ncbi.nlm.nih.gov/entrez/) and full papers (especially the Material and Methods sections) where available from PubMed Central (http://www.pubmedcentral.gov/) (Spasic et al. 2007).
A fragment of the NMR ontology in Protégé, the class/subclass relationship (taxonomy) on the left and the description of the term/class on the right hand side (including definition and administrative metadata). Top-level classes, imported from OBI or BFO, are displayed in the hierarchy with a lighter shade
The initial source of terms for the CV is Rubtsov et al. (2007). As stated before, the minimal reporting requirements and the format are both work in progress conducted by other MSI WGs, therefore, the ontology for the NMR sub-component should not be considered complete at this stage. The NMR.owl has also served as a test bed to evaluate the BFO top-level ontology as well as technical issues such as the OWL-import, cross ontology referencing mechanism, modularisation, constraint inheritance and the usage of RA and RU metadata annotation properties in Protégé. Overall, we can say that this experience has been an excellent use case to practice our working strategy and collaboration with the larger OBI group.
Concluding remarks
Every effort will be made to meet the group goals in a timely fashion, although the MSI OWG members are geographically distributed and central funds do not exits for the MSI WGs activities. One of the major bottlenecks in building bio-ontologies is the lack of a unified methodology and tools for collaborative development, making large collaborative endeavours more challenging (Castro et al. 2006). The MSI OWG and the OBI WG pose scenarios in which domain experts are geographically widespread and the structure of the ontology is constantly evolving. Consequently face-to-face workshops have proved to be the most efficient way to significantly advance the project. In addition, the sociological barriers involved in these kinds of large-scale collaborations can be far more challenging and extensive liaison is necessary between communities. Managing this process of consensus building from start to finish requires ample time, resources and expertise. The time invested to identify commonalities and synergies with other projects, such as OBI, is often limited due to a lack of resources. The massively collaborative nature of the ontology undertaking requires frequent face-to-face workshops to create the optimal conditions for building of consensus. Teleconferences and web meetings are also used, but these are generally short and are not an ideal mechanism for efficient collaborative development; rarely are they as effective as direct interactions established at face-to-face workshops. Unfortunately it is very difficult to hold such workshops without central funds; such funds having previously been difficult to obtain in competition with more traditional scientific projects. In the special issue of the journal OMICS (Field and Sansone 2006) twenty invited manuscripts describe different standardisation initiatives focusing on both the successes and pitfalls encountered, and lessons learned. This issue also includes a special call for action for further recognition of the importance of global omics standardisation activities (Brooksbank and Quackenbush 2006), where the authors eloquently describe the Herculean efforts that are often accomplished ‘on the side’ and without formal funding, simply because the lack of standardisation is an unacceptable state of affairs for omics researchers and is repeatedly proving to be a significant bottleneck in the collection, querying, processing, and sharing of data.
References
Allen, F. H., Barnard, J. M., Cook, A. P. F., & Hall, S. R. (1995). The Molecular Information File (MIF): Core Specifications of a New Standard Format for Chemical Data. Journal of Chemical Information and Computer Sciences, 35, 412–427.
Blake, J. A., & Bult, C. J. (2006). Beyond the data deluge: data integration and bio-ontologies. Journal of Biomedical Informatics, 39, 314–320.
Bodenreider, O. (2004). The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research, 32, D267–D270.
Bodenreider, O., & Stevens, R. (2006). Bio-ontologies: current trends and future directions. Briefings in Bioinformatics, 7, 256–274.
Brooksbank, C., & Quackenbush, J. (2006). Data standards: a call to action. Omics, 10, 94–99.
Castle, A. L., Fiehn, O., Kaddurah-Daouk, R., & Lindon, J. C. (2006). Metabolomics Standards Workshop and the development of international standards for reporting metabolomics experimental results. Briefings in Bioinformatics, 7, 159–165.
Castro, A. G., Rocca-Serra, P., Stevens, R., Taylor, C., Nashar, K., Ragan, M. A., & Sansone, S. A. (2006). The use of concept maps during knowledge elicitation in ontology development processes-the nutrigenomics use case. BMC Bioinformatics, 7, 267.
Cimino, J. J., & Zhu, X. (2006). The practical impact of ontologies on biomedical informatics. Methods of Information in Medicine, 45(Suppl 1), 124–135.
de Matos, P., Ennis, M., Darsow, M., Guedj, M., Degtyarenko, K., & Apweiler, R. (2006). ChEBI—Chemical Entities of Biological Interest. Nucleic Acids Research.
Fiehn, O., Kristal, B., van Ommen, B., Sumner, L. W., Sansone, S. A., Taylor, C., Hardy, N., & Kaddurah-Daouk, R. (2006). Establishing reporting standards for metabolomic and metabonomic studies: a call for participation. Omics, 10, 158–163.
Fiehn, O., Sumner, L. W., Rhee, S. Y., Ward, J., Dickerson, J., Lange, B. M., Lane, G., Roessner, U., Last, R., & Nikolau, B. (2007). Minimum reporting standards for plant biology context information in metabolomics studies. Metabolomics, 3, this issue.
Field, D., & Sansone, S. A. (2006). A special issue on data standards. Omics, 10, 84–93.
Griffin, J. L., Nicholls, A. W., Daykin, C. A., Heald, S., Keun, H. C., Schuppe-Koistinen, I., Griffiths, J. R., Cheng, L. L., Rocca-Serra, P., Rubtsov, D. V., & Robertson, D. (2007). Standard reporting requirements for biological samples in metabolomics experiments: mammalian / in vivo experiments. Metabolomics, 3, this issue.
Hardy, N. W., & Taylor, C. F. (2007). A roadmap for the establishment of standard data exchange structures for metabolomics. Metabolomics, 3, this issue.
Hermjakob, H. (2006). The HUPO Proteomics Standards Initiative—Overcoming the Fragmentation of Proteomics Data. Proteomics, 6, 34–38.
Jenkins, H., Hardy, N., Beckmann, M., Draper, J., Smith, A. R., Taylor, J., Fiehn, O., Goodacre, R., Bino, R. J., Hall, R., Kopka, J., Lane, G. A., Lange, B. M., Liu, J. R., Mendes, P., Nikolau, B. J., Oliver, S. G., Paton, N. W., Rhee, S., Roessner-Tunali, U., Saito, K., Smedsgaard, J., Sumner, L. W., Wang, T., Walsh, S., Wurtele, E. S., & Kell, D. B. (2004). A proposed framework for the description of plant metabolomics experiments and their results. Nature Biotechnology, 22, 1601–1606.
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., Katayama, T., Araki, M., & Hirakawa, M. (2006). From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Research, 34, D354–D357.
Lan, N., Montelione, G. T., & Gerstein, M. (2003). Ontologies for proteomics: towards a systematic definition of structure and function that scales to the genome level. Current Opinion in Chemical Biology, 7, 44–54.
Lindon, J. C., Nicholson, J. K., Holmes, E., Keun, H. C., Craig, A., Pearce, J. T., Bruce, S. J., Hardy, N., Sansone, S. A., Antti, H., Jonsson, P., Daykin, C., Navarange, M., Beger, R. D., Verheij, E. R., Amberg, A., Baunsgaard, D., Cantor, G. H., Lehman-McKeeman, L., Earll, M., Wold, S., Johansson, E., Haselden, J. N., Kramer, K., Thomas, C., Lindberg, J., Schuppe-Koistinen, I., Wilson, I. D., Reily, M. D., Robertson, D. G., Senn, H., Krotzky, A., Kochhar, S., Powell, J., van der Ouderaa, F., Plumb, R., Schaefer, H., & Spraul, M. (2005). Summary recommendations for standardization and reporting of metabolic analyses. Nature Biotechnology, 23, 833–838.
Morrison, N., Bearden, D., Bundy, J. G., Collette, T., Currie, F., Davey, M. P., Haigh, N. S., Hancock, D., Jones, O. A. H., Rochfort, S., Sansone, S-A., Stys, D., Teng, Q., Field, D., & Viant, M. R. (2007). Standard reporting requirements for biological samples in metabolomics experiments: environmental context. Metabolomics, 3, this issue.
Noy, N. F., Crubezy, M., Fergerson, R. W., Knublauch, H., Tu, S. W., Vendetti, J., & Musen, M. A. (2003) Protege-2000: An Open-source Ontology-development and Knowledge-acquisition Environment. Proc AMIA Symp, 953.
Quackenbush, J. (2004). Data standards for ‘omics science. Nature Biotechnology, 22, 613–614.
Rickard, K., Mejino, J., Martin, R. J., Agoncillo, A., & Rosse, C. (2004). Problems and solutions with integrating terminologies into evolving knowledge bases. Medinfo, 11, 420–424.
Rosse, C., Kumar, A., Mejino, J. L. Jr., Cook, D. L., Detwiler, L. T., & Smith, B. (2005) A strategy for improving and integrating biomedical ontologies. AMIA Annu Symp Proc, 639–643.
Rubin, D. L., Lewis, S. E., Mungall, C. J., Misra, S., Westerfield, M., Ashburner, M., Sim, I., Chute, C. G., Solbrig, H., Storey, M. A., Smith, B., Day-Richter, J., Noy, N. F., & Musen, M. A. (2006). National center for biomedical ontology: Advancing biomedicine through structured organization of scientific knowledge. Omics, 10, 185–198.
Rubtsov, D. V., Jenkins, H., Ludwig, C., Easton, J., Viant, M. R., Gunther, U., Griffin, J. L., & Hardy, N. (2007). Proposed reporting requirements for the description of NMR-based metabolomics experiments. Metabolomics, 3, this issue.
Sansone, S. A., Rocca-Serra, P., Tong, W., Fostel, J., Morrison, N., & Jones, A. R. (2006). A strategy capitalizing on synergies: The Reporting Structure for Biological Investigation (RSBI) working group. Omics, 10, 164–171.
Schober, D., Kusnierczyk, W., Lewis, S., Lomax, J., Members of the MSI, PSI Ontology Working Groups, Mungall, S., Rocca-Serra, P., Smith B., & Sansone, S.-A. (2007). Towards naming conventions for use in controlled vocabulary and ontology engineering. In Proceedings of the Bio-Ontologies Workshop, ISMB/ECCB, Vienna http://bio-ontologies.org.uk/download/Bio-Ontologies2007.pdf, pp. 29–32.
Schulze-Kremer, S. (1998). Ontologies for molecular biology. Pac Symp Biocomput, 695–706.
Schulze-Kremer, S. (2002) Ontologies for molecular biology and bioinformatics. In Silico Biology, 2, 179–193.
Shulaev, V. (2006). Metabolomics technology and bioinformatics. Briefings in Bioinformatics, 7, 128–139.
Smith, B., Ceusters, W., Klagges, B., Kohler, J., Kumar, A., Lomax, J., Mungall, C., Neuhaus, F., Rector, A. L., & Rosse, C. (2005). Relations in biomedical ontologies. Genome Biology, 6, R46.
Smith, B., Kusnierczyk, W., Schober, D., & Ceusters, W. (2006). Towards a Reference Terminology for Ontology Research and Development in the Biomedical Domain. KR-MED 2006.
Smith, B., Ashburner, M., Rosse, C., Bard, J., Bug, W., Ceusters, W., Goldberg, L., Eilbeck, K., Ireland, A., Mungall, C., the OBI Consortium, Leontis, N., Rocca-Serra, P., Ruttenberg, A., Sansone, S-A., Shah, N., Whetzel, P. L., Lewis, S. The OBO Foundry: Coordinated Evolution of Ontologies to Support Biomedical Data Integration. Nature Biotechnology (under review).
Soldatova, L. N., & King, R. D. (2006). An ontology of scientific experiments. Journal of the Royal Society Interface.
Spasic, I., Dunn, W. B., Velarde, G., Tseng, A., Jenkins, H., Hardy, N., Oliver, S. G., & Kell, D. B. (2006). MeMo: A hybrid SQL/XML approach to metabolomic data management for functional genomics. BMC Bioinformatics, 7, 281.
Spasic, I., Schober, D., Sansone, S-A., Rebholz-Schuhmann, D., Kell, D. B., Paton, N., & MSI Ontology Working Group Members (2007). Facilitating the development of controlled vocabularies for metabolomics with text mining. In Proceedings of the Bio-Ontologies Workshop, ISMB/ECCB, Vienna, http://bio-ontologies.org.uk/download/Bio-Ontologies2007.pdf, pp. 45–48.
Stevens, R., Bodenreider, O., & Lussier, Y. A. (2006). Semantic webs for life sciences. Pacific Symposium on Biocomputing, 112–115.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., Fan, T. W-M., Fiehn, O., Goodacre, R., Griffin, J. L., Hankemeier, T., Hardy, N., Harnly, J., Higashi, R., Kopka, J., Lane, A. N., Lindon, J. C., Marriott, P., Nicholls, A. W., Reily, M. D., Thaden, J. J., & Viant, M. R. (2007). Proposed minimum reporting standards for chemical analysis. Metabolomics, 3, this issue.
Supekar, K., & Musen, M. (2005). Ontology metadata to support the building of a library of biomedical ontologies. AMIA Annual Symposium Proceedings, 1127.
Taylor, C. F., Hermjakob, H., Julian, R. K. Jr., Garavelli, J. S., Aebersold, R., & Apweiler, R. (2006). The work of the Human Proteome Organisation’s Proteomics Standards Initiative (HUPO PSI). Omics, 10, 145–151.
van der Werf, M. J., Takors, R., Smedsgaard, J., Nielsen, J., Ferenci, T., Portais, J. C., Wittmann, C., Hooks, M., Tomassini, A., Oldiges, M., Fostel, J., & Sauer, U. (2007). Standard reporting requirements for biological samples in metabolomics experiments: microbial and in vitro biology experiments. Metabolomics, 3, this issue.
Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, M., Ulrich, E. L., Markley, J. L., Ionides, J., & Laue, E. D. (2005). The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins, 59, 687–696.
Whetzel, P. L., Brinkman, R. R., Causton, H. C., Fan, L., Field, D., Fostel, J., Fragoso, G., Gray, T., Heiskanen, M., Hernandez-Boussard, T., Morrison, N., Parkinson, H., Rocca-Serra, P., Sansone, S. A., Schober, D., Smith, B., Stevens, R., Stoeckert, C. J. Jr., Taylor, C., White, J., & Wood, A. (2006). Development of FuGO: an ontology for functional genomics investigations. Omics, 10, 199–204.
Wishart, D. S., Tzur, D., Knox, C., Eisner, R., Guo, A. C., Young, N., Cheng, D., Jewell, K., Arndt, D., Sawhney, S., Fung, C., Nikolai, L., Lewis, M., Coutouly, M. A., Forsythe, I., Tang, P., Shrivastava, S., Jeroncic, K., Stothard, P., Amegbey, G., Block, D., Hau, D. D., Wagner, J., Miniaci, J., Clements, M., Gebremedhin, M., Guo, N., Zhang, Y., Duggan, G. E., Macinnis, G. D., Weljie, A. M., Dowlatabadi, R., Bamforth, F., Clive, D., Greiner, R., Li, L., Marrie, T., Sykes, B. D., Vogel, H. J., & Querengesser, L. (2007). HMDB: the Human Metabolome Database. Nucleic Acids Research, 35, D521–D526.
Acknowledgements
The authors are members of the MSI Ontology WG; Susanna-Assunta Sansone is the current acting chair and Daniel Schober is the post-doctoral ontologist assisting the WG with the developmental phases. We kindly acknowledge the MSI Oversight Committee, the other MSI WGs chairs and members, the OBI working group, the OBO Foundry leaders and the Ontogenesis Networks members for their contributions in fruitful discussions. We also gratefully thank the BBSRC e-Science Development Fund (BB/D524283/1 and BB/E025080/1, to Susanna-Assunta Sansone), the BBSRC MeT-RO project (MET20483, to Helen Jenkins), the BBSRC/EPSRC “The Manchester Centre for Integrative Systems Biology” (to Irena Spasic), the EU Network of Excellence NuGO (NoE 503630, to Philippe Rocca-Serra) and the EU Network of Excellence Semantic Interoperability and Data Mining in Biomedicine (NoE 507505, supporting Daniel Schober and Irena Spasic exchange visits).
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
See the MSI Ontology Working Group website for a complete list of members and contributors. Web URL: http://msi-workgroups.sf.net
Rights and permissions
About this article
Cite this article
Sansone, SA., Schober, D., Atherton, H.J. et al. Metabolomics standards initiative: ontology working group work in progress. Metabolomics 3, 249–256 (2007). https://doi.org/10.1007/s11306-007-0069-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-007-0069-z