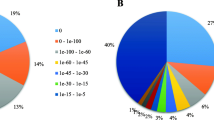
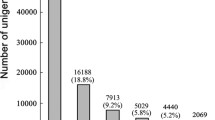
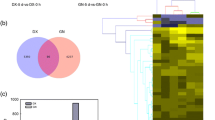
Abstract
Eucalyptus nitens (H. Deane & Maiden) is a fast-growing species used principally for pulpwood and solid-wood production. Due to its cold tolerance, it is preferred over other Eucalyptus species at high elevations. To get a deeper insight in the molecular mechanisms of cold acclimation, the transcriptome profiling by RNA-Seq in plants of E. nitens under cold acclimation and deacclimation process was compared in order to identify differentially expressed genes (DEGs). Transcriptomes from control, cold acclimated to chilling temperature, cold acclimated at freezing temperature, and deacclimation condition were compared using Eucalyptus grandis as reference genome. The differential expression analysis allowed the identification of a total of 1600 DEGs out of which 1088 and 1071 were identified in response to cold acclimation and deacclimation, respectively. The gene ontology analysis revealed that DEGs were significantly enriched in response to stimulus, response to abiotic stimulus, membrane, catalytic activity, and cell periphery. Furthermore, the biochemical pathways analysis revealed a large number of DEGs represented in the biosynthesis of phenylpropanoids, specifically flavonoid biosynthesis likely to support ROS scarvening, genes related to photosynthesis, genes that take part in glycolysis/gluconeogenesis related to starch biosynthesis pathway, and genes represented in carotenoid biosynthesis pathway suggesting a role in the regulation of ABA synthesis, which has been previously involved in stress tolerance. A total of 115 DEGs corresponding to transcription factors were identified, being the most represented families AP2, MYB, and WRKY. Expression of six DEGs was validated using qRT-PCR that further supported the in silico results. The present study provides a comprehensive view of global gene expression and revealed valuable information about the dynamic and complex nature of gene expression occurring during cold acclimation and deacclimation process in E. nitens.





Similar content being viewed by others
References
Agarwal M, Hao Y, Kapoor A, Dong C-H, Fujii H, Zheng X, Zhu J-K (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Aguayo P et al (2016) Overexpression of an SKn-dehydrin gene from Eucalyptus globulus and Eucalyptus nitens enhances tolerance to freezing stress in Arabidopsis. Trees-Struct Funct 30:1785–1797. doi:10.1007/s00468-016-1410-9
Anders S, Pyl PT, Huber W (2015) A Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–10.
Arora R, Wisniewski ME (1994) Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica [L.] Batsch) (II. A 60-kilodalton bark protein in cold-acclimated tissues of peach is heat stable and related to the dehydrin family of proteins). Plant Physiol 105:95–101
Barrero-Gil J, Salinas J (2013) Post-translational regulation of cold acclimation response. Plant Sci 205-206:48–54. doi:10.1016/j.plantsci.2013.01.008
Beck EH, Fettig S, Knake C, Hartig K, Bhattarai T (2007) Specific and unspecific responses of plants to cold and drought stress. J Biosci 32:501–510
Brunetti C, Di Ferdinando M, Fini A, Pollastri S, Tattini M (2013) Flavonoids as antioxidants and developmental regulators: relative significance in plants and humans. Int J Mol Sci 14:3540–3555
Cansev A, Gulen H, Eris A (2009) Cold-hardiness of olive (Olea europaea L.) cultivars in cold-acclimated and non-acclimated stages: seasonal alteration of antioxidative enzymes and dehydrin-like proteins. J Agric Sci 147:51–61
Cao PB, Azar S, SanClemente H, Mounet F, Dunand C, Marque G, Marque C, Teulières C (2015) Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: an intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE 10(4):e0121041
Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289
Cassan-Wang H et al (2012) Reference genes for high-throughput quantitative reverse transcription–PCR analysis of gene expression in organs and tissues of Eucalyptus grown in various environmental conditions. Plant Cell Physiol 53:2101–2116
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11:113–116
Chawade A, Bräutigam M, Lindlöf A, Olsson O, Olsson B (2007) Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors. BMC Genomics 8:304
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10:281
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA)—Gene Regulatory Mechanisms 1819:120–128
Chen L, Zhong H-y, Kuang J-f, Li J-g, Lu W-j, Chen J-y (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234:377–390
Chen S et al (2014) Performance comparison between rapid sequencing platforms for ultra-low coverage sequencing strategy. PLoS One:9
Close D, Beadle C, Battaglia M (2004) Foliar anthocyanin accumulation may be a useful indicator of hardiness in eucalypt seedlings. For Ecol Manag 198:169–181
Costa e Silva F et al (2009) Acclimation to short-term low temperatures in two Eucalyptus globulus clones with contrasting drought resistance. Tree Physiol 29:77–86. doi:10.1093/treephys/tpn002
Dugald CC, Chris LB, Philip HB, Greg KH (2000) Cold-induced photoinhibition affects establishment of Eucalyptus nitens (Deane and Maiden) maiden and Eucalyptus globulus Labill. Trees. doi:10.1007/s004680000070
El Kayal W, Navarro M, Marque G, Keller G, Marque C, Teulieres C (2006) Expression profile of CBF-like transcriptional factor genes from Eucalyptus in response to cold. J Exp Bot 57:2455–2469. doi:10.1093/jxb/erl019
Fernández M, Troncoso V, Valenzuela S (2015) Transcriptome profile in response to frost tolerance in Eucalyptus globules. Plant Mol Biol Rep 33:1472–1485
Fernández M, Valenzuela Águila S, Arora R, Chen K (2012a) Isolation and characterization of three cold acclimation-responsive dehydrin genes from Eucalyptus globulus. Tree Genet Genomes 8:149–162. doi:10.1007/s11295-011-0429-8
Fernández M, Valenzuela S, Barraza H, Latorre J, Neira V (2012b) Photoperiod, temperature and water deficit differentially regulate the expression of four dehydrin genes from Eucalyptus globulus. Trees 26:1483–1493
Fernández M, Villarroel C, Balbontín C, Valenzuela S (2010) Validation of reference genes for real-time qRT-PCR normalization during cold acclimation in Eucalyptus globulus. Trees 24:1109–1116. doi:10.1007/s00468-010-0483-0
Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54:767–781
Gonzalez-Aguero M, Garcia-Rojas M, Di Genova A, Correa J, Maass A, Orellana A, Hinrichsen P (2013) Identification of two putative reference genes from grapevine suitable for gene expression analysis in berry and related tissues derived from RNA-Seq data. BMC Genomics 14:878
Guy CL (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Biol 41:187–223
Hara M, Terashima S, Kuboi T (2001) Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J Plant Physiol 158:1333–1339
Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61:1041–1052
Huang DW, Sherman BT, Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13
Huang DW, Sherman BT, Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Huang L, Yan H, Jiang X, Yin G, Zhang X, Qi X, et al. (2014) Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PloS One 9:e93724.
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118
Janská A, Maršík P, Zelenková S, Ovesná J (2010) Cold stress and acclimation—what is important for metabolic adjustment? Plant Biol 12:395–405
Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J (2016) WRKY transcription factors in plant responses to stresses Journal of Integrative Plant Biology. doi:10.1111/jipb.12513.
Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kasuga J, Hashidoko Y, Nishioka A, Yoshiba M, Arakawa K, Fujikawa S (2008) Deep supercooling xylem parenchyma cells of katsura tree (Cercidiphyllum japonicum) contain flavonol glycosides exhibiting high anti-ice nucleation activity. Plant Cell Environ 31:1335–1348
Keller G, Cao PB, San Clemente H, El Kayal W, Marque C, Teulières C (2013) Transcript profiling combined with functional annotation of 2,662 ESTs provides a molecular picture of Eucalyptus gunnii cold acclimation. Trees 27:1713–1735
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi:10.1186/gb-2013-14-4-r36
Külheim C, Yeoh SH, Maintz J, Foley WJ, Moran GF (2009) Comparative SNP diversity among four Eucalyptus species for genes from secondary metabolite biosynthetic pathways. BMC Genomics 10:452
Lee JM, Roche JR, Donaghy DJ, Thrush A, Sathish P (2010) Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol Biol 11:8
Leek JT, Taub MA, Rasgon JL (2012) A statistical approach to selecting and confirming validation targets in-omics experiments. BMC bioinformatics 13:1
Li C-F et al (2015) Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis). BMC Genomics 16:560
Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET (2013) Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 200:457–472
Liu H-c, Charng Y-y (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163:276–290
Liu Y, Jiang Y, Lan J, Zou Y, Gao J (2014) Comparative transcriptomic analysis of the response to cold acclimation in Eucalyptus dunnii. PLoS One 9(11):e113091
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Mangul S, Caciula A, Al Seesi S, Brinza D, Mӑndoiu I, Zelikovsky A (2014) Transcriptome assembly and quantification from ion torrent RNA-Seq data. BMC Genomics 15:S7
Mangwanda R, Myburg AA, Naidoo S (2015) Transcriptome and hormone profiling reveals Eucalyptus grandis defence responses against Chrysoporthe austroafricana. BMC Genomics 16:319
Marian CO, Eris A, Krebs SL, Arora R (2004) Environmental regulation of a 25 kDa dehydrin in relation to Rhododendron cold acclimation. J Am Soc Hortic Sci 129:354–359
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Miura K, Furumoto T (2013) Cold signaling and cold response in plants. Int J Mol Sci 14:5312–5337
Mizrachi E, Hefer CA, Ranik M, Joubert F, Myburg AA (2010) De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genomics 11:681
Monroy AF, Sangwan V, Dhindsa RS (1998) Low temperature signal transduction during cold acclimation: protein phosphatase 2A as an early target for cold-inactivation. Plant J 13:653–660
Moraga PS, Escobar R, Valenzuela SA (2006) Resistance to freezing in three Eucalyptus globulus Labill subspecies. Electron J Biotechnol 9(3):310–314
Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12:3788–3796
Moura JCMS, Araújo P, dos Brito MS, Souza UR, Viana JOF, Mazzafera P (2012) Validation of reference genes from Eucalyptus spp. under different stress conditions. BMC research notes 5:634
Mowla SB, Cuypers A, Driscoll SP, Kiddle G, Thomson J, Foyer CH, Theodoulou FL (2006) Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J 48:743–756
Navarrete-Campos D, Bravo LA, Rubilar RA, Emhart V, Sanhueza R (2013) Drought effects on water use efficiency, freezing tolerance and survival of Eucalyptus globulus and Eucalyptus globulus × nitens cuttings. New For 44:119–134
Navarro M, Ayax C, Martinez Y, Laur J, El Kayal W, Marque C, Teulieres C (2011) Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnol J 9:50–63
Navarro M, Marque G, Ayax C, Keller G, Borges JP, Marque C, Teulieres C (2009) Complementary regulation of four Eucalyptus CBF genes under various cold conditions. J Exp Bot 60:2713–2724. doi:10.1093/jxb/erp129
Nguyen HC et al. (2016) Special trends in CBF and DREB2 groups in Eucalyptus gunnii vs Eucalyptus grandis suggest that CBF are master players in the trade-off between growth and stress resistance Physiologia plantarum. doi:10.1111/ppl.12529
Oates CN, Külheim C, Myburg AA, Slippers B, Naidoo S (2015) The transcriptome and terpene profile of Eucalyptus grandis reveals mechanisms of defense against the insect pest, Leptocybe invasa. Plant Cell Physiol 56:1418–1428
Oquist G, Huner NP (2003) Photosynthesis of overwintering evergreen plants. Annu Rev Plant Biol 54:329–355. doi:10.1146/annurev.arplant.54.072402.115741
Pearce RS (2001) Plant freezing and damage. Ann Bot 87:417–424
Powell LE (1987) The hormonal control of bud and seed dormancy in woody plants. In: Plant hormones and their role in plant growth and development. (Davies, P.J., ed.). Dordrecht: Martinus-Nijhoff, pp. 539–552
Quail MA et al (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341
Rajkumar AP et al (2015) Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC Genomics 16:1
Ray DL, Johnson JC (2014) Validation of reference genes for gene expression analysis in olive (Olea europaea) mesocarp tissue by quantitative real-time RT-PCR. BMC research notes 7:304
Raymond C, Harwood C, Owen J (1986) A conductivity method for screening populations of eucalypts for frost damage and frost tolerance. Aust J Bot 34:377–393. doi:10.1071/BT9860377
Read J, Busby J (1990) Comparative responses to temperature of the major canopy species of Tasmanian cool temperate rain-forest and their ecological significance. II. Net photosynthesis and climate analysis. Aust J Bot 38:185–205
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi:10.1093/bioinformatics/btp616
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants the. Plant J 23:319–327
Salleh FM et al (2012) A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ 35:418–429
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi:10.1093/bioinformatics/btr026
Singh KB, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Söderman E, Mattsson J, Engström P (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J 10:375–381
Steyn W, Wand S, Holcroft D, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155:349–361
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Strickler SR, Bombarely A, Mueller LA (2012) Designing a transcriptome next-generation sequencing project for a nonmodel plant species1. Am J Bot 99:257–266
Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21:2213–2223
Team RC (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
Teklemariam TA, Blake TJ (2004) Phenylalanine ammonia-lyase-induced freezing tolerance in jack pine (Pinus banksiana) seedlings treated with low, ambient levels of ultraviolet-B radiation. Physiol Plant 122:244–253
Thavamanikumar S, Southerton S, Thumma B (2014) RNA-Seq using two populations reveals genes and alleles controlling wood traits and growth in Eucalyptus nitens. PLoS One 9:e101104
Thomashow MF (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 118:1–8. doi:10.1104/pp.118.1.1
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Biol 50:571–599
Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577. doi:10.1104/pp.110.161794
Thumma BR, Sharma N, Southerton SG (2012) Transcriptome sequencing of Eucalyptus camaldulensis seedlings subjected to water stress reveals functional single nucleotide polymorphisms and genes under selection. BMC Genomics 13:364
Tibbits W, Boomsma D, Jarvis S (1997) Distribution, biology, genetics, and improvement programs for Eucalyptus globulus and E. nitens around the world. In: White T, Huber D, Powell G (eds), Proceedings of the 24th biennial southern tree improvement conference, June 9–12 1997. Southern Tree Improvement Committee, Orlando, Florida, pp. 1–15
Tibbits W, Reid J (1987) Frost resistance in Eucalyptus nitens (Deane & Maiden) Maiden: physiological aspects of hardiness. Aust J Bot 35:235–250
Tran LSP et al (2007) Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 49:46–63
Turnbull J, Eldridge K (1983) The natural environment of Eucalyptus as the basis for selecting frost resistant species. In: Proceedings of IUFRO/AFOCEL symposium on frost resistant Eucalyptus. (26th – 30th September 1983, Bordeaux, France). p. 43–62.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi:10.1093/nar/gks596
Van Buskirk HA, Thomashow MF (2006) Arabidopsis transcription factors regulating cold acclimation. Physiol Plant 126:72–80
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034
Villar E, Klopp C, Noirot C, Novaes E, Kirst M, Plomion C, Gion J-M (2011) RNA-Seq reveals genotype-specific molecular responses to water deficit in Eucalyptus. BMC Genomics 12:538
Vining KJ, Contreras RN, Ranik M, Strauss SH (2012) Genetic methods for mitigating invasiveness of woody ornamental plants: research needs and opportunities. HortScience. 47:1210–1216.
Wang L et al (2014) Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol 14:103
Wang X-C et al (2013) Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:415
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1) England:57–63. doi:10.1038/nrg2484
Weiser C (1970) Cold resistance and injury in woody plants knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science 169:1269–1278
Welling A, Palva ET (2006) Molecular control of cold acclimation in trees. Physiol Plant 127:167–181
Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J 8:749–771
Wisniewski M, Close TJ, Artlip T, Arora R (1996) Seasonal patterns of dehydrins and 70-kDa heat-shock proteins in bark tissues of eight species of woody plants. Physiol Plant 96:496–505
Wisniewski M, Nassuth A, Teulieres C, Marque C, Rowland J, Cao PB, Brown A (2014) Genomics of cold hardiness in woody plants. Crit Rev Plant Sci 33:92–124
Wisniewski M, Norelli J, Bassett C, Artlip T, Macarisin D (2011) Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233:971–983
Wright IJ, Reich P, Westoby M (2001) Strategy shifts in leaf physiology, structure and nutrient content between species of high-and low-rainfall and high-and low-nutrient habitats. Funct Ecol 15:423–434
Yakovlev IA, Asante DK, Fossdal CG, Partanen J, Junttila O, Johnsen Ø (2008) Dehydrins expression related to timing of bud burst in Norway spruce. Planta 228:459–472
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi:10.1186/1471-2105-13-134
Zhao Z, Tan L, Dang C, Zhang H, Wu Q, An L (2012) Deep-sequencing transcriptome analysis of chilling tolerance mechanisms of a subnival alpine plant. Chorispora bungeana BMC plant biology 12:222
Zheng C, Zhao L, Wang Y, Shen J, Zhang Y, Jia S, et al. (2015) Integrated RNA-Seq and sRNASeq analysis identifies chilling and freezing responsive key molecular players and pathways in tea plant (Camellia sinensis). PLoS One 10(4):e0125031
Acknowledgements
Financial support came from Fondecyt Iniciación 11121559 and Genómica Forestal S.A. We would like to thank Conicyt for scholarship to CL and MFB.
Author’s contributions
VE provided plant material. CL and MFB performed growth chamber experiments. JGL performed the RNA-Seq data analysis. CL and MFB performed the validation of reference genes and CA-responsive genes by qRT-PCR. JGL, CL, and MF drafted the manuscript. MF initiated, designed, and led the project. SV contributed to experimental design. All authors contributed to manuscript preparation and editing. All authors read and approved the final manuscript.
Data archiving statement
The short-read sequences data have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession SRP066573. The Bioproject ID related to this paper is PRJNA303180.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.B. Neale
Rights and permissions
About this article
Cite this article
Gaete-Loyola, J., Lagos, C., Beltrán, M.F. et al. Transcriptome profiling of Eucalyptus nitens reveals deeper insight into the molecular mechanism of cold acclimation and deacclimation process. Tree Genetics & Genomes 13, 37 (2017). https://doi.org/10.1007/s11295-017-1121-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1121-4